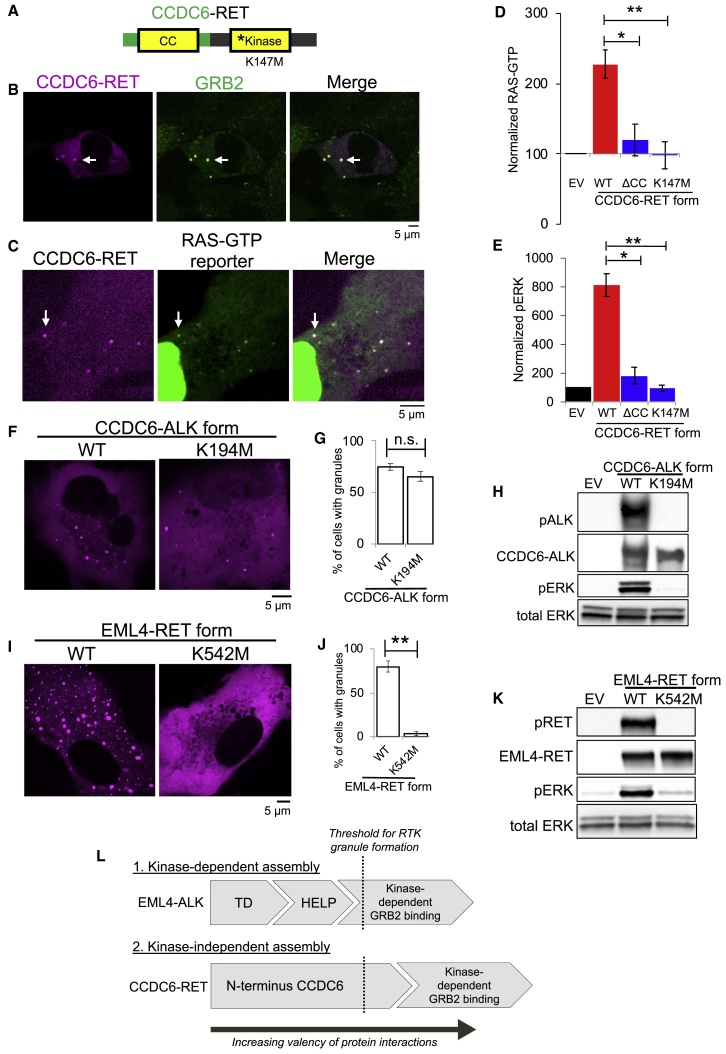
Figure 6.
Cytoplasmic granule formation is a general mechanism for RTK-mediated RAS/MAPK pathway activation in cancer
(A) Domain structure schematic of CCDC6-RET.
(B) Live-cell imaging of mTagBFP2::CCDC6-RET in Beas2B cells with endogenous mNG2-tagging of GRB2.
(C) Live-cell imaging of Beas2B cells expressing mTagBFP2::CCDC6-RET and RAS-GTP reporter.
(D and E) Quantification of endogenous RAS-GTP levels (D) and ERK phosphorylation (E) by western blotting in 293T cells expressing EV, CCDC6-RET WT, or mutant forms. n = 4.
(F and G) Live-cell imaging of mEGFP-tagged CCDC6-ALK WT or kinase-deficient K194M mutant in Beas2B cells. Quantification of % cells with granules (6 or greater) from 75 total cells, n = 3.
(H) Western blotting upon expression of EV, CCDC6-ALK WT or K194M mutant in 293T cells.
(I and J) Live-cell imaging of mEGFP-tagged EML4-RET WT or kinase-deficient K542M mutant in Beas2B cells. Quantification of % cells with granules from 75 total cells, n = 3.
(K) Western blotting upon expression of EV, EML4-RET WT, or K542M mutant in 293T cells.
(L) Simplified threshold model for RTK protein granule formation based on cumulative valency of protein interactions contributed by N-terminal RTK fusion partner and kinase-dependent GRB2 binding in each protein assembly context.
For (B) and (C), arrows indicate a representative CCDC6-RET cytoplasmic protein granule with local enrichment of GRB2 (B) or RAS-GTP reporter (C) (multiple non-highlighted granules also show colocalization). For all panels, microscopy images representative of at least 75 cells analyzed over 3 replicates. Error bars represent ±SEM, ∗∗p < 0.01, ∗p < 0.05. n.s., non-significant comparison, one-way ANOVA with post hoc Tukey’s HSD test (D and E) or paired t test (G and J).
See also Figure S6.