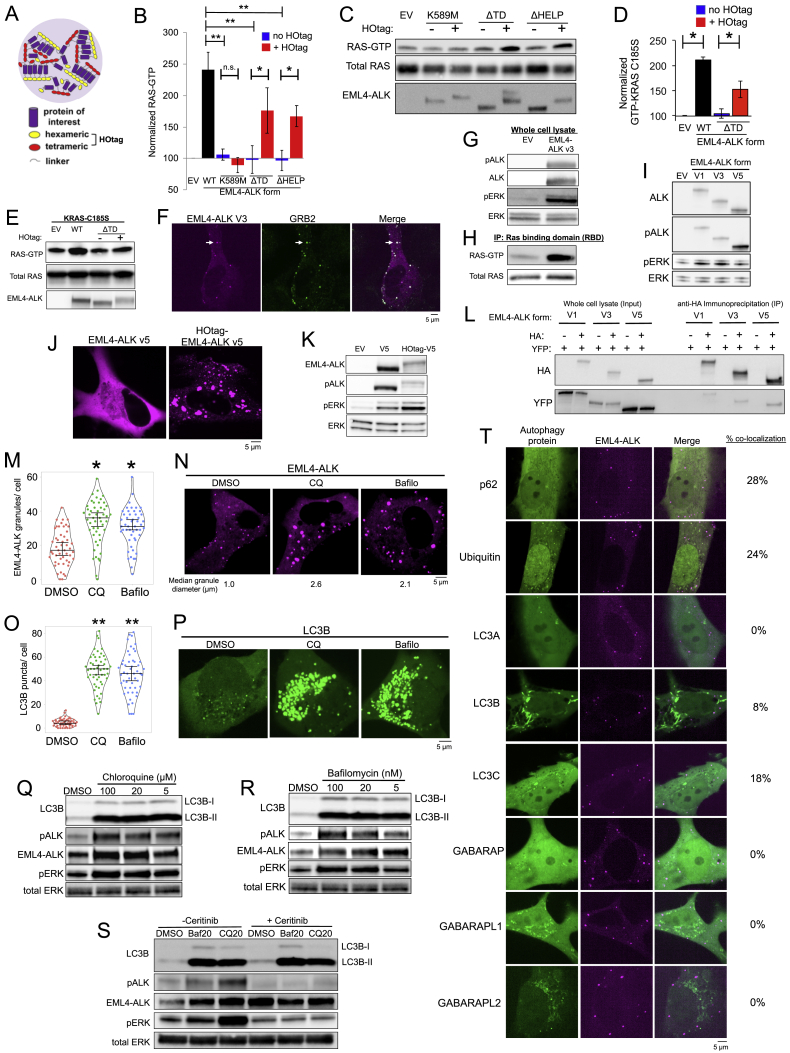
Figure S5.
Higher-order granule formation is critical for RAS/MAPK signaling, related to Figure 5
(A) Forced clustering of proteins achieved by N-terminal hexameric and C-terminal tetrameric tags that form higher-order clustered assemblies upon expression in cells.
(B, C) Quantification of endogenous RAS-GTP levels and representative western blots from 293T cells expressing an empty vector (EV), EML4-ALK wild-type (WT), or the diffusely cytosolic EML4-ALK mutants (kinase deficient K589M, ΔTD, or ΔHELP) +/– forced clustering (HOtag). EML4-ALK K589M, ΔTD and ΔHELP mutants (blue bars) display significantly reduced RAS-GTP levels compared to wild-type EML4-ALK (black bar), ∗∗ denotes p < 0.01 by one-way ANOVA with post hoc Tukey’s HSD test. Forced clustering (HOtag, red bars) of EML4-ALK ΔTD and ΔHELP mutants, but not EML4-ALK K589M, significantly increases RAS-GTP levels compared to the respective non-clustered EML4-ALK mutants (blue bars), ∗ denotes p < 0.05, n.s. denotes non-significant comparison, paired t test. RAS-GTP levels normalized to total RAS protein levels and then internally normalized to EML4-ALK mutant expression levels. n = 4. Error bars represent ± SEM.
(D, E) Stable expression of cytosolic KRAS mutant (KRAS C185S) in 293T cells, followed by transfection of empty vector (EV), EML4-ALK wild-type (WT) or diffusely cytosolic EML4-ALK ΔTD mutant ± forced clustering (HOtag). Levels of GTP-bound KRAS C185S were normalized to total KRAS C185S protein levels and then internally normalized to expression level of EML4-ALK form. Western blot images representative of n = 4 independent experiments. Error bars represent ± SEM, ∗ denotes p < 0.05 by paired t test.
(F) Live-cell confocal imaging of mTagBFP2::EML4-ALK variant 3 expressed in human epithelial cell line Beas2B with endogenous mNG2-tagging of GRB2. White arrows indicate a representative EML4-ALK variant 3 cytoplasmic protein granule with local enrichment of GRB2 (multiple non-highlighted granules also show colocalization between EML4-ALK variant 3 and GRB2). Images are representative of at least 25 analyzed cells in 3 independent experiments.
(G, H) Western blot analysis upon expression of empty vector (EV) or EML4-ALK variant 3 in 293T cells to assess levels of EML4-ALK activation (phosphorylation), ERK activation, and in immunoprecipitation (IP) panel (H), levels of RAS activation (GTP-bound RAS). Representative images from at least 4 independent experiments.
(I) Western blot analysis upon expression of empty vector (EV) or EML4-ALK variants 1, 3, or 5 in 293T cells to assess levels of ERK activation. Images are representative of at least 4 independent experiments and pERK levels are quantified in Main Figure 5J.
(J) Live-cell confocal imaging of YFP::EML4-ALK variant 5 ± forced clustering (HOtag) in human epithelial cell line Beas2B. Images are representative of at least 20 analyzed cells in 3 independent experiments.
(K) Western blot analysis upon expression of empty vector (EV) or EML4-ALK variant 5 ± HOtag in 293T cells. Images are representative of at least 4 independent experiments.
(L) YFP- and HA-tagged versions of EML4-ALK variants 1, 3, and 5 were expressed in 293T cells as indicated. Western blot analysis of input and immunoprecipitation (IP) with anti-HA beads. Images are representative of 3 independent experiments.
(M, N) Violin plot of EML4-ALK granule number and representative live-cell confocal images of Beas2B cells expressing YFP::EML4-ALK treated with DMSO, 20 μM chloroquine (CQ), or 20 nM bafilomycin (Bafilo) for 24 hours. At least 50 cells were analyzed per condition over 3 independent experiments. Median diameter of granules calculated from analysis of 20 cells. ∗ denotes p < 0.05 by one-way ANOVA with post hoc Tukey’s HSD test.
(O, P) Violin plot of number of LC3B puncta and representative live-cell confocal images of Beas2B cells expressing mEGFP::LC3B treated with DMSO, 20 μM chloroquine (CQ), or 20 nM bafilomycin (Bafilo) for 24 hours. At least 50 cells were analyzed per condition over 3 independent experiments. ∗∗ denotes p < 0.01 by one-way ANOVA with post hoc Tukey’s HSD test.
(Q, R, S) Western blot analysis in EML4-ALK expressing H3122 cancer cell line treated with listed doses of chloroquine or bafilomycin for 24 hours. 500 nM ceritinib was added 1 hour prior to harvesting lysates (S). Images are representative of 4 independent experiments.
(T) Live-cell confocal images of Beas2B cells expressing mTagBFP2::EML4-ALK and mEGFP-tagged LC3/ATG8 family proteins. Images are representative of 3 independent experiments. Percent colocalization calculated from at least 60 cells in a total of 3 independent experiments. Standard error for percent colocalization: p62 (2.1), Ubiquitin (4.0), LC3B (0.9), and LC3C (2.6).