Figure 2. Top lncRNA screen hits in HeLa cells are located in the HPV18 integration locus.
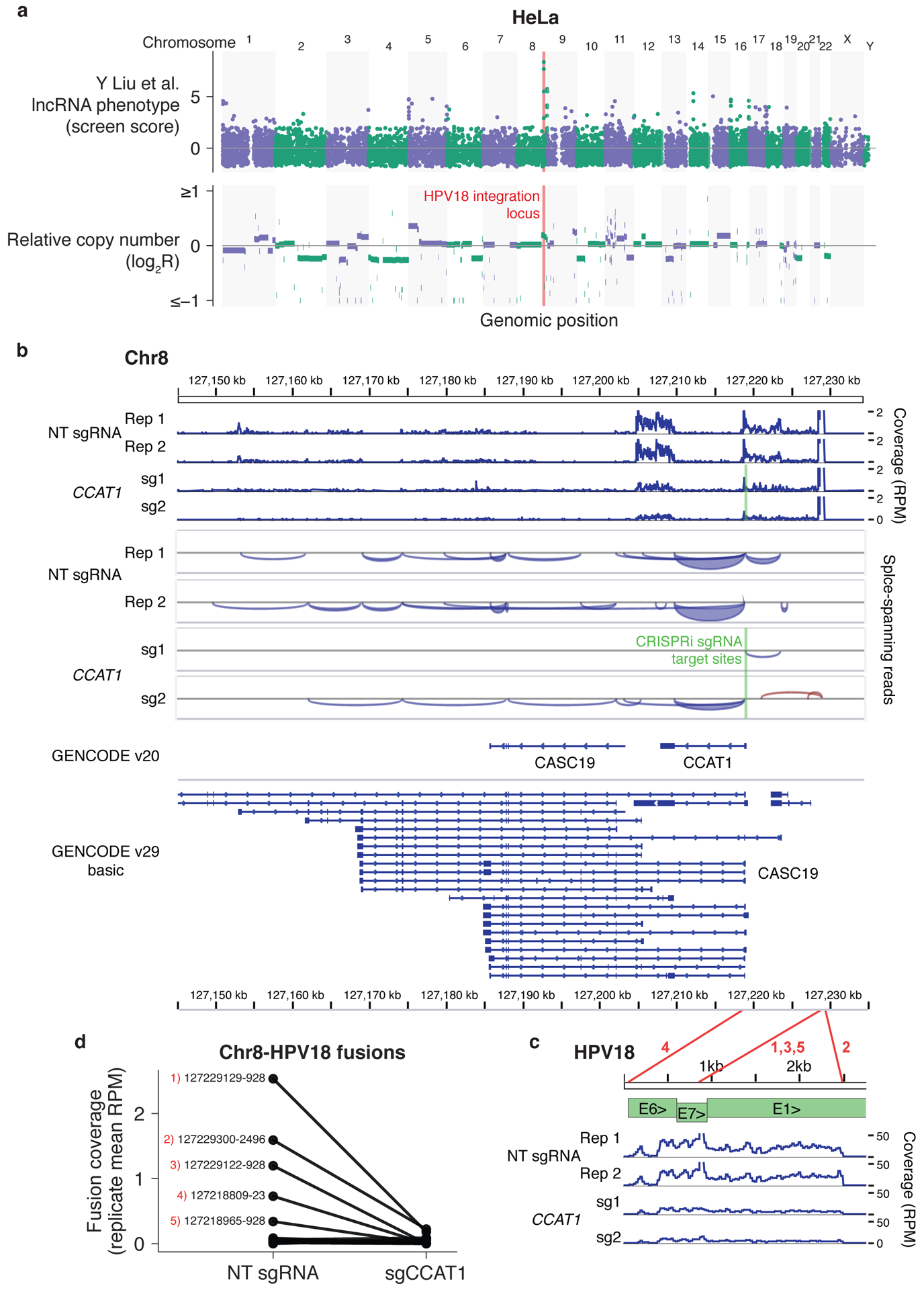
(a) HeLa lncRNA screen scores (top) and relative copy number (bottom) across the genome. (b) RNA-seq coverage across the chromosome 8 locus indicated in (a) with and without CCAT1 knockdown with CRISPRi. (c) RNA-seq coverage across the HPV18 genome using published RNA-seq datasets5 aligned to the hg38 and HPV18 genome sequences with TopHat-Fusion without annotation. Coverage in reads per million (RPM) was calculated with IGVtools count, and coverage, splice spanning reads, and two GENCODE annotations were visualized in IGV. HPV18 gene structure adapted from Adey et al.8 Red lines correspond to fusion sites reported by TopHat-Fusion and numbers correspond to the labeled fusion sites in (d). (d) RNAseq coverage of chromosome 8–HPV18 fusion sites, averaged across two independent sgRNAs. Coverage was determined by the number of fusion-spanning reads reported by TopHat-Fusion normalized to total aligned reads. The top five fusion sites by coverage are labeled with the respective chromosome 8 and HPV18 genome coordinates.
