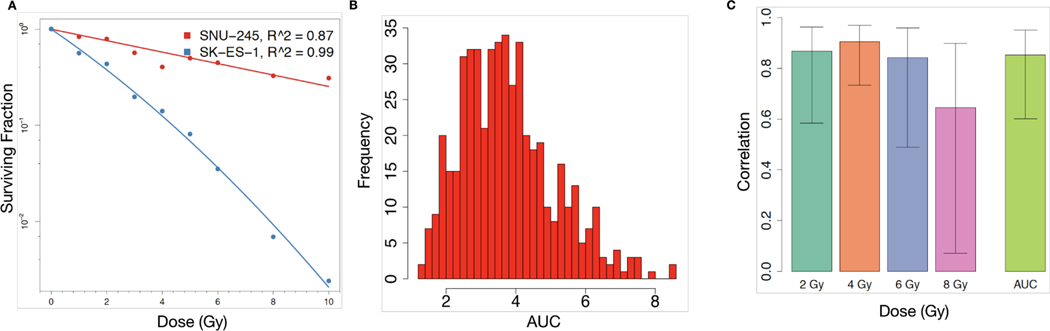
Figure 2: Fitting of dose-response data to the LQ model and concordance of radiation response across asays.
(A) LQ model fit using RadioGx on the SNU-245 cholangiocarcinoma cell line (red) and SK-ES-1 Ewing sarcoma cell line (blue). The LQ model describes the fraction of cells predicted to survive (y-axis) a uniform radiation dose (x-axis) and is characterized by α and β components for each cell line. For SNU-245 and SK-ES-1, α = 0.14 (Gy−1), β (Gy−2) = 0 and α = 0.45 (Gy−1), β = 0.02 (Gy−2), respectively. Solid curves indicate the model fit and points denote experimental data (Yard et al., 2016). (B) Histogram of AUC values calculated using the computeAUC function in RadioGx. (C) Correlation (Pearson R with standard deviation) of radioresponse results produced by the 9-day viability assay and the standard clonogenic assay according to the following indicators: SF2, SF4, SF6, SF8, and AUC. Primary data were obtained from Yard et al..