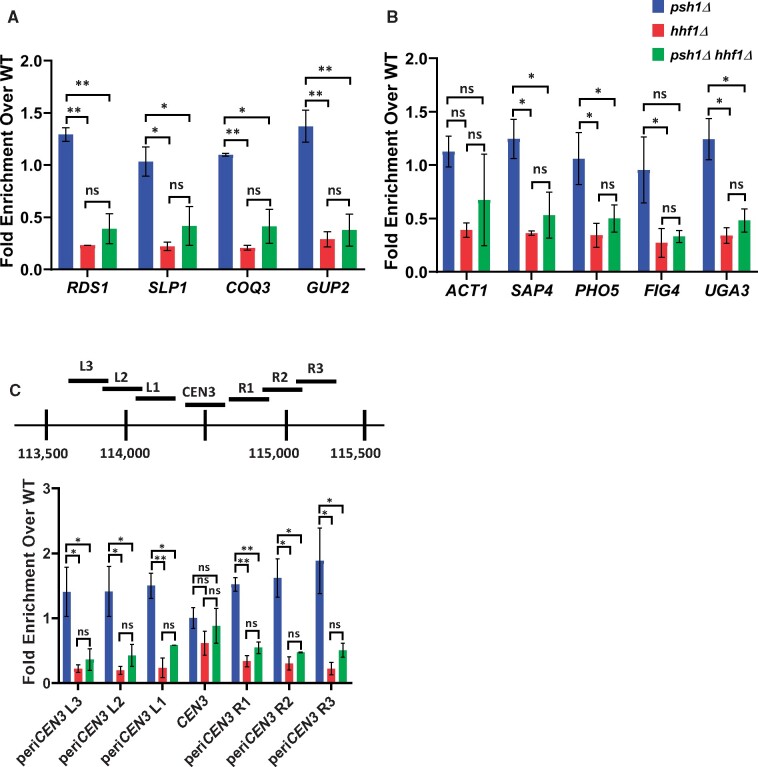
Figure 3.
Deletion of the HHF1 allele reduces enrichment of Cse4 at peri-centromeric and noncentromeric regions. (A–C) ChIP-qPCR was performed on chromatin lysate from wild-type (YMB9804), psh1Δ (YMB10479), hhf1Δ (YMB10937), and psh1Δ hhf1Δ (YMB10822) strains transiently overexpressing GAL1-6His-3HA-CSE4 (pMB1458). Enrichment of 6His-3HA-Cse4 is shown as a fold over wild-type. Displayed are the mean of two independent experiments. Error bars represent standard deviation of the mean. **P < 0.0099, *P < 0.09, ns, not significant. (A and B) Levels of Cse4 enrichment are reduced at noncentromeric regions when HHF1 is deleted. qPCR at A: ACT1, SAP4, RDS1, SLP1, and PHO5 and B: FIG4, COQ3, GUP2, and UGA3. (C) Cse4 enrichment is reduced at peri-centromeric, but not the core centromere, regions in hhf1Δ strains. Top: A diagram of the peri-centromere and centromere of Chromosome III analyzed by ChIP-qPCR. Horizontal lines represent the regions amplified. Bottom: Enrichment of 6His-3HA-Cse4 at the core centromere and the left and right peri-centromeric regions on Chromosome III.