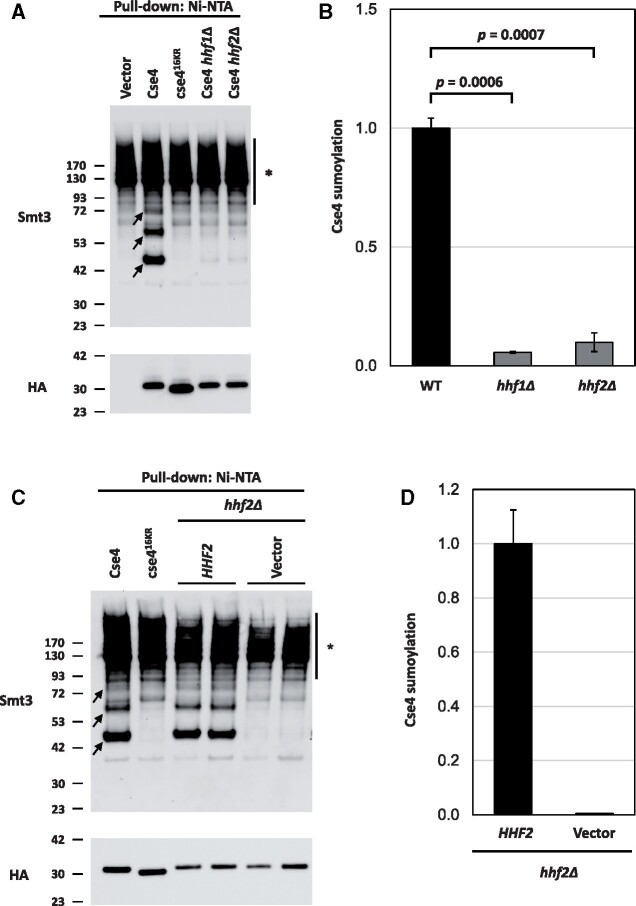
Figure 5.
Histone H4 contributes to the sumoylation of Cse4. (A) Levels of sumoylated Cse4 are decreased in histone H4 deletion strains. Sumoylation levels were assayed on wild-type (BY4741) strains transformed with empty vector (pYES2), pGAL-8His-HA-CSE4 (pMB1345), or pGAL-8His-HA-cse416KR (pMB1344) and hhf1Δ (YMB10766) and hhf2Δ (YMB10767) strains transformed with pGAL-8His-HA-Cse4 (pMB1345). Sumoylated and nonmodified Cse4 were detected using cell lysates that were incubated with Ni-NTA beads followed by western blot analysis with antibodies against Smt3 and HA (Cse4), respectively. Arrows indicate the three high molecular weight bands that represent sumoylated Cse4. Asterisk indicates nonspecific sumoylated proteins that bind to beads. (B) Quantification of relative levels of sumoylated Cse4 in histone H4 deletion strains. Levels of sumoylated Cse4 were normalized to nonmodified Cse4 probed against HA in the pull-down sample. Statistical significance from two independent experiments was assessed by one-way ANOVA (P = 0.0004) followed by Tukey post-test (all pairwise comparisons of means). Error bars indicate average deviation from the mean. (C) The Cse4 sumoylation defect in a hhf2Δ strain is linked to the HHF2 allele. Sumoylation levels were determined from lysates from a hhf2Δ (YMB10767) strain with pGAL-8His-HA-CSE4 (pMB1345) transformed with vector (pRS425) or HHF2 (pMB1929) as described in (A). Arrows indicate the three high molecular weight bands that represent sumoylated Cse4. Asterisk indicates nonspecific sumoylated proteins that bind to beads. (D) Quantification of relative levels of sumoylated Cse4. Relative levels of sumoylated Cse4 were normalized to nonmodified Cse4 probed against HA in the pull-down sample. Error bars indicate average deviation from the mean from two biological replicates.