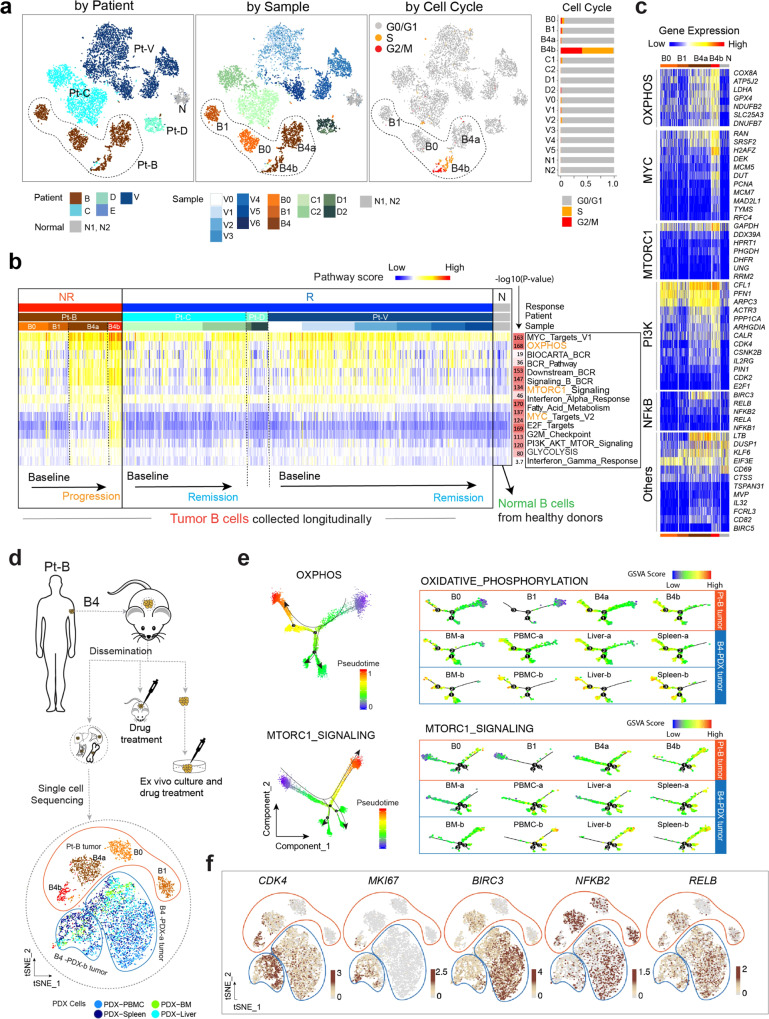
Fig. 2. The transcriptomic heterogeneity and evolution of cancer hallmarks is associated with therapeutic resistance.
a Color-coded t-SNE plots of the malignant B cells. Color-coded cell representation by subject (Responders: C, D, V; non-responders: B; and healthy donors: N1, N2), sample collection time, and by cell cycle stage. b Transcriptomic heterogeneity and evolution of cancer hallmarks associated with ibrutinib resistance. From the Molecular Signature Database (MSigDB), 50 hallmark cancer gene sets were downloaded, and a pathway activity score was calculated for each single cell. The top 13 cancer hallmark pathways upregulated in the progressive sample are shown. c Heatmap representation of differentially expressed genes (representative ones) from five selected pathways across cell sub-populations B0, B1, B4a, and B4b from patient B compared to normal samples (N). d Schematic view of the establishment of B4-derived PDX model and experimental strategy of sample collection for scRNA-seq analysis. e Developmental trajectories representation of malignant cell populations from patient B (B0, B1, B4a, and B4b) and B4-derived PDX tumors along inferred pseudotime by Monocle2. Each point corresponds to a single cell; all points are color-coded according to the inferred pseudotime. Monocle 2 was run with default parameters on the hallmark gene sets (OXPHOS and mTORC1 signaling) downloaded from MSigDB. f tSNE Plots featuring CKD4, MKI67, BIRC3, NFKB2 and RELB genes expression in cells from patient B tumor and B4-derived PDX tumors.