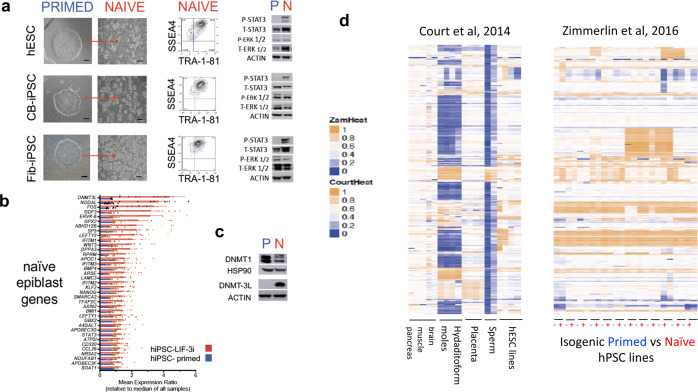
Fig. 5. Chemical reversion of conventional primed hPSC with a tankyrase/PARP inhibitor promotes stable rewiring to a human preimplantation epiblast-like state with intact epigenomic imprints.
The TIRN method is a two-step culture system109,111 comprised of one brief adaptation step of conventional hPSC with LIF and five small molecules (LIF-5i) that includes XAV939 (tankyrase/PARP inhibition), CHiR99021 (GSK3β inhibition), PD0325901 (MEK inhibition), forskolin (adenylate cyclase activation), and purmorphamine (Hedgehog signaling activation); followed by continuous, stable culture in only LIF/XAV939/CHIR99021/PD0325901 (LIF-3i) for at least 30 passages. This tankyrase/PARP inhibitor-mediated modification of the classical LIF-2i method has been validated to stably revert over 30 hPSC lines, and is independent of genetic donor background109,111. a (Left panels) Three representative hPSC lines109,110 (RUES1 hESC, cord blood (CB)-hiPSC 6.2, and fibroblast (fib)-hiPSC C.2 are shown in their starting primed (i.e., E8 medium; PRIMED) conditions, and 6–10 passages post culture in continuous TIRN-reverted (NAÏVE) conditions109,111. Monolayer bFGF-dependent hPSC colonies in primed conditions became tolerant to bulk single-cell passaging and acquired a typical dome-shape morphology following TIRN reversion. (Middle panels) TIRN-hPSC retained strong expression of TRA-1–81 and SSEA4 surface antigens by flow cytometry. (Right panels) Western blot analyses in these three primed (P) and TIRN (N) lines demonstrated that TIRN-hPSC acquired de novo expression of phosphorylated STAT3 and reduced ERK1/2 phosphorylation. b TIRN-hPSC uniformly shifted their transcriptomes toward a pre-implantation naïve epiblast identity. Mean expression ratios of naïve epiblast-specific genes from Illumina gene arrays from isogenic primed-to-naïve-reverted pairs of genetically independent hPSC (n = 9 lines)109. c Western blot analysis of primed (P) and TIRN (N) CB-hiPSC (E5C3). TIRN-CB-hiPSC markedly upregulated naïve epiblast-specific DNMT3L whilst maintaining DNMT1 protein expressions following TIRN reversion109,110. d Infinium CpG methylation heatmaps of genomic Imprinted promoter regions of a panel of primed and TIRN-reverted isogenic hPSC samples revealed that TIRN-hPSC retains normal somatic epigenomic imprint configurations, similar to their isogenic primed states. Heatmaps compare previously published Zambidis lab109 methylation data alongside published Court et al.121 data of identical imprinted genomic regulatory regions as controls. Zambidis lab methylation beta values were subset to exact imprinted regions provided by Court et al.121 (methylation beta values: 0—completely hypomethylated probe; 1—completely methylated probe). Matching Infinium probes are sorted by chromosomal location and arranged into their adjacent primed (−) and naïve (+) hPSC isogenic pairs. The heatmaps of the same imprinted regions from Court et al.121 include abnormal androgenetic hydatidiform mole, normal human tissues (e.g., brain, muscle, placenta, and sperm), and primed hESC lines.