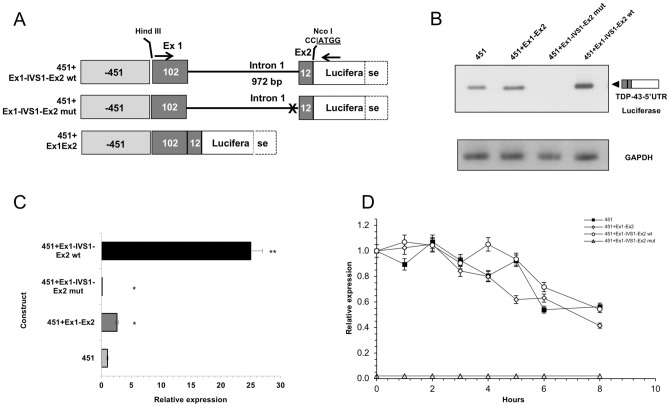
Figure 8.
Effects of 5′UTR and intron 1 on the transcriptional activity of the human TARDBP promoter. (A) Diagrams of the 5′UTR TDP-43 constructs generated using the pGL-451 vector. In the 451 + Ex1-IVS1-Ex2 wt construct, the 1086 bp genomic region of the human TARDBP gene spanning the 5′UTR, including Exon 1 (102 bp), Intron 1 (972 bp) to the first 12 nt of Exon 2) was (Hind III-Nco I) cloned in between the 451nt-promoter and the firefly luciferase ORF (Luciferase). In the 451 + Ex1-IVS1-Ex2 mut construct, the 3′ splicing site of Intron 1 was deleted. In the 451 + Ex1Ex2 construct, the fragment encompassing Exon 1 and Exon 2 (without Intron 1) was cloned in the 451 vector. (B) RT-PCR of the 5′UTR TDP-43 mRNA species (upper panel). Amplification of GAPDH (lower panel) was used as the endogenous control in the quantitative analysis of RT-PCR. (C) Quantitation of the 5′UTR TDP-43 mRNA species by Real Time PCR. GAPDH was used to confirm normalization of total RNA levels. Co-transfected renilla orf was used to normalize the luciferase qPCR. The 451 construct was used as reference (= 1). (D) Effects of transcriptional inhibition on mRNA stability. After 36 h post-transfection of the luciferase (451 + Ex1-IVS1-Ex2 wt and 451 + Ex1-IVS1-Ex2 mut) constructs, SH-SY5Y cells were treated with actinomycin D 5 µg/ml. The relative levels of the indicated mRNAs (wt and mut) were assessed at the designated time points, following shutoff of transcription using qRT-PCR to determine mRNA half-lives. The average half-lives are reported with SD from two independent experiments. Significance values refer to comparisons against t = 0 h, wt: *p < 0.05 (one way ANOVA with Tukey test). The mean values for 451 + Ex1-IVS1-Ex2 mut construct were negligible and did not differ to a statistically significant extent (p ≥ 0.05).