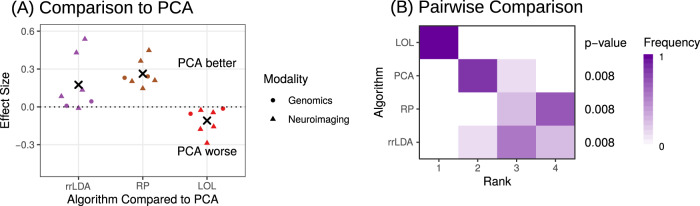
Fig. 3. Comparing various dimensionality reduction algorithms on two real datasets: neuroimaging and genomics.
A Beeswarm plots show the classification performance of each technique with respect to PCA at the optimal number of embedding dimensions, the number of embedding dimensions with the lowest misclassification rate. Performance is measured by the effect size, defined as κ(LDA∘ PCA) − κ(LDA∘embed), where κ is Cohen’s Kappa, and embed is one of the embedding techniques compared to PCA. Each point indicates the performance of PCA relative the other technique on a single dataset, and the sample size-weighted average effect is indicated by the black “x.” LOL always outperforms PCA and all other techniques. B Frequency histograms of the relative ranks of each of the embedding techniques on each dataset after classification, where a 1 indicates the best relative classification performance and a 4 indicates the worst relative classification performance, after embedding with the technique indicated. Projecting first with LOL provides a significant improvement over competing strategies (Wilcoxon signed-rank test, n = 7, p value = 0.008) on all benchmark problems.