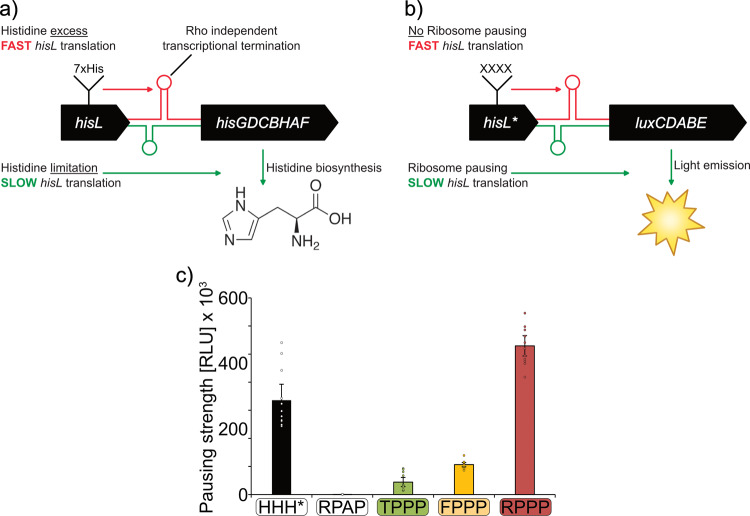
Fig. 4. The His-pausing system for in vivo measurement of pausing strength.
a Architecture of the histidine biosynthesis operon in E. coli. In its native state, the histidine biosynthesis gene cluster (hisGDCBHAF) is regulated by the His-leader peptide (hisL). This peptide contains seven consecutive histidines. At high histidine/histidyl-tRNA levels, translation efficiently proceeds through the His-leader peptide, resulting in the formation of an attenuator stem loop (red) that prevents transcription of the downstream genes. At low histidine and histidyl-tRNA levels translation is slowed down allowing for transcription and translation of the structural genes and synthesis of histidine (green). b Architecture of the His-pausing operon. An engineered His-leader peptide (hisL*) precedes the structural genes of the lux operon (luxCDABE). Here, His1 through His4 are exchanged by artificial sequence motifs (XXXX). In case of non-consecutive proline motifs (e.g., RPAP) there is no pausing, resulting in the formation of an attenuator stem loop (red) that prevents transcription of the downstream genes and low light emission. In the presence of motifs that contain consecutive prolines (e.g., RPPP) translation is slowed down allowing for transcription and translation of the structural genes and thus increased light emission (green). c Maximal luminescence emission at PP-motifs with increasing pausing strength. HisL*_Lux operons carrying a stop codon at the position corresponding to His4 (HHH*), non-consecutive (RPAP) or consecutive prolines of varying known pausing strength at the hisL* position (Weak: TPPP; green. Intermediate: FPPP; yellow. Strong: RPPP; red) were chromosomally integrated in E. coli BW25113 and tested for maximal luminescence emission. Threonine, phenylalanine, and arginine were encoded by ACC, TTT, and CGC, respectively. CCG was used as proline codon in all constructs. n = 12, Error bars indicate 95% confidence intervals.