FIGURE 3.
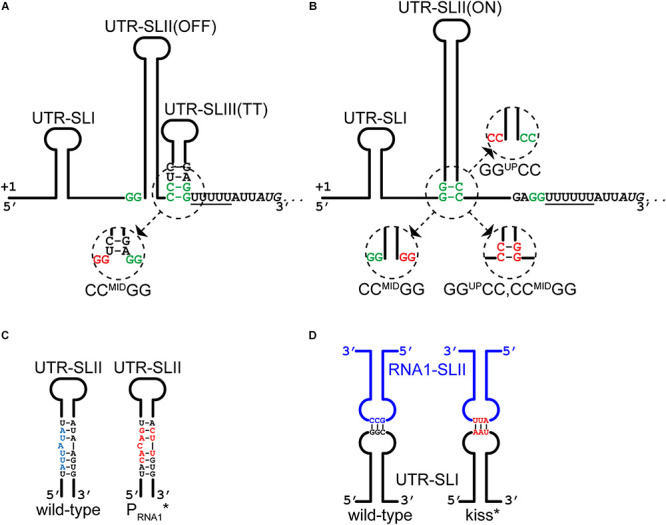
Mutations that alters RNA1-repA mRNA interactions. (A) The CCMIDGG mutation (red nucleotides in insert) prevents the formation of the putative OFF-structure by shortening the duplex of the stem and liberating some of the nucleotides of the ribosome binding site. (B) The CCMIDGG mutation (red nucleotides in insert) removes two base-pairs from the foot of the 46 base-pair UTR-SLII(ON) stem, in addition to preventing the putative OFF-structure from forming (A). The GGUPCC mutation (red nucleotides in insert) also weakens the UTR-SLII stem, which presumably shifts the equilibrium toward formation of UTR-SLIII(TT). The GGUPCC,CCMIDGG mutation (red nucleotides in insert) allows the full length of the UTR-SLII(ON) stem to form, and at the same time weakens/prevents the UTR-SLIII(TT). (C) The PRNA1* mutations in the –10 region of the RNA1 promoter (red nucleotides) do not disrupt base-pairing within the UTR-SLII (wild-type in blue). See also Supplementary Figure 2 for full nucleotide sequence. (D) The kiss* mutations (in red) weaken the base-base interactions between RNA1-SLII and UTR-SLI. See also Supplementary Figure 2 for full nucleotide sequence.
