FIGURE 6.
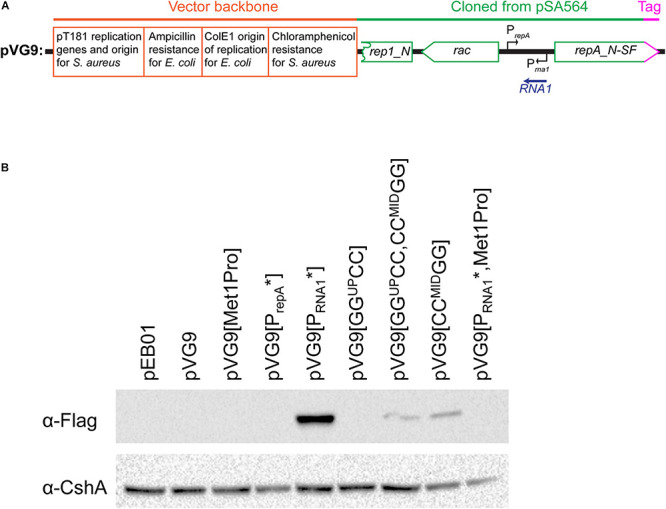
Mutating the RNA1 promoter permits the detection of RepA by Western blotting.(A) Schematic linear map of the pVG9 construct. The orange section is from the vector backbone, which includes a pT181 replication cassette (with replication genes and origin) for replication in S. aureus, a ColE1 origin of replication for E. coli, an ampicillin resistance cassette for E. coli and a chloramphenicol resistance cassette for S. aureus. The green section was cloned from pSA564, and includes (from left to right) a truncated rep1_N gene, the rac gene, the RNA1 gene, and the repA_N gene. The latter is translationally fused to a streptavidine-FLAG tag (pink). The location of the rac promoter is unknown. Further details about the pSA564 insert and the backbone cloning vector can be found in Supplementary Figure 1. (B) RepA is undetectable under normal circumstances (pVG9) but becomes easily detectable when the RNA1 promoter is mutated (pVG9[PRNA1∗]). An additional mutation of the RepA start codon abolishes detection (pVG9[PRNA1∗, Met1Pro]), thus confirming the specificity of the antibody. Mutations that prevent formation of the putative OFF-structure (pVG9[GGUPCC,CCMIDGG] and pVG9[CCMIDGG]) also permits the detection of RepA. In the construct “PrepA∗” the -10 box of the repA promoter was mutated from TAATAT to TAATGG, in “Met1Pro” the AUG start codon of RepA was mutated to CCG. In the pVG9 constructs the RepA was C-terminally fused to a streptavidine-FLAG tag, and was detected using anti-FLAG antibodies. Antibodies against the CshA protein were used as loading control.
