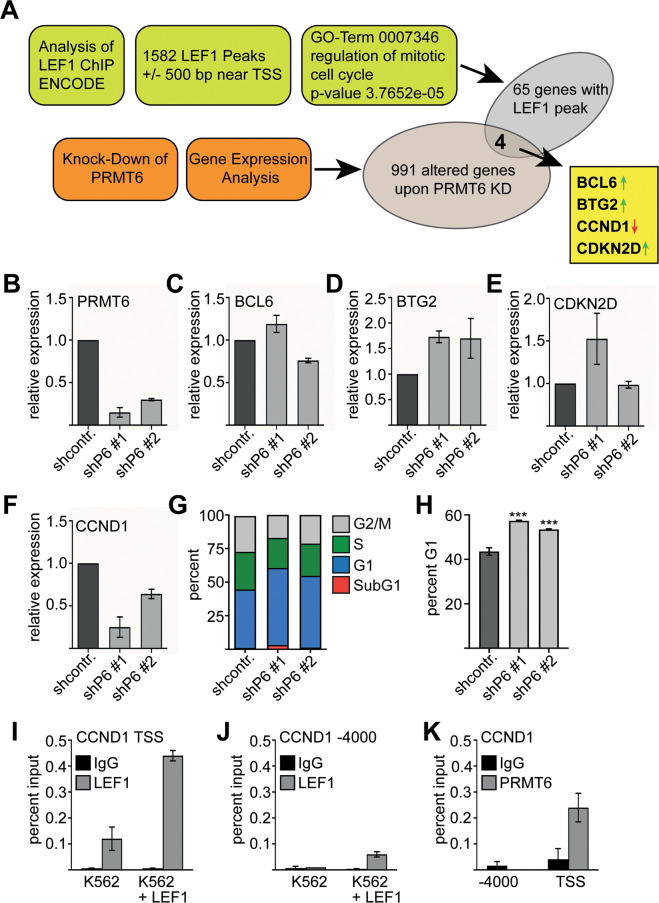
Fig. 4. Combined ChIP-seq and RNA transcriptome analysis reveals CCND1 as a direct LEF1/PRMT6 target.
A Evaluation of LEF1 ChIP Encode data in K562 cells and GO-term analysis revealed 65 genes regulating the mitotic cell cycle, which are bound by LEF1. Expression analysis identified 991 differentially expressed genes upon knockdown of PRMT6 in K562 cells. Of these, four genes are cell cycle associated LEF1 targets, BCL6, BTG2, CCND1, and CDKN2D. The arrows indicate upregulation or downregulation upon PRMT6 knockdown. B mRNA expression analysis of two different shRNA constructs against PRMT6 (shP6). GAPDH expression was used for normalization. C–F The four identified cell cycle associated LEF1/PRMT6 targets were re-analysed by quantitative real-time PCR 7 days after shPRMT6 transduction. Error bars represent the standard deviation from at least three independent experiments. G Cell cycle analysis was performed five days after PRMT6 knockdown in K562 cells. H Percentage of cells within the G1 phase of the cell cycle increased upon PRMT6 knockdown in K562 cells. I, J ChIP assay shows that LEF1 is bound close to the CCND1 transcriptional start site (TSS). This binding is increased upon LEF1 over expression. The negative control (−4000 bp from the TSS) is displayed in J. K ChIP-assay shows that PRMT6 is bound close to the CCND1 transcriptional start site (TSS), but not to the −4000 region. ChIP were performed with an anti-LEF1 and anti-PRMT6 antibody, respectively. The P-values were calculated using Student’s t-test from at least three independent measurements. *P < 0.05; **P < 0.01; ***P < 0.001.