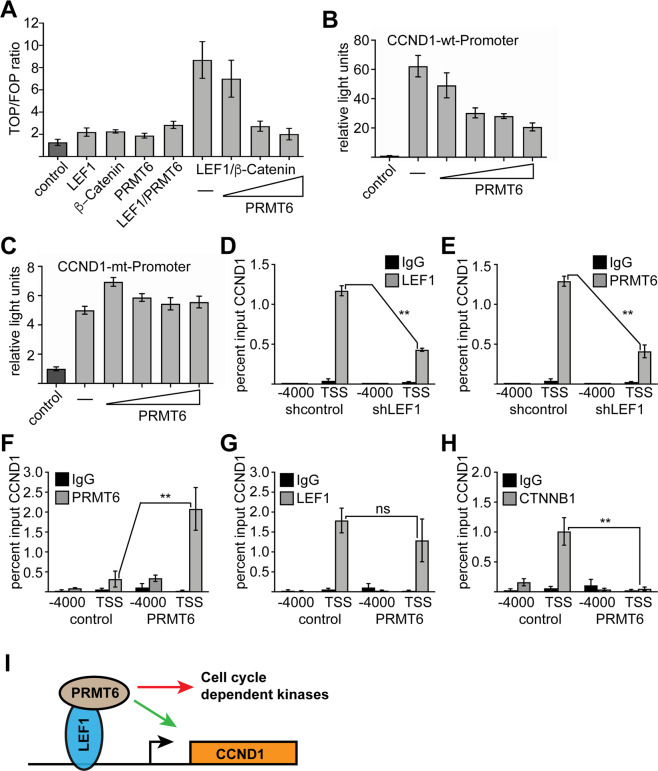
Fig. 6. Interdependence of LEF1 and PRMT6 on CCND1.
A PRMT6 TOP-Flash assay. The M50 Super 8x TOP-Flash11 was co-transfected with LEF1, PRMT6 and N89-(constantly active) β-Catenin12 into HEK293T cells. The negative control M51 Super 8× FOP-Flash was used for normalization. Co-transfection of LEF1 and N89-ß-Catenin leads to the highest TOP/FOP ratio. Increasing PRMT6 amount decreased the activating effect of β-Catenin. B, C CCND1 luciferase assay. The bars represent the mean with standard deviation of two independent experiments, each measured in technical duplicates. D, E ChIP assay was performed seven days upon knockdown of LEF1 in K562 cells. Binding of LEF1 and PRMT6 to the CCND1 promoter was reduced upon LEF1 knockdown. F ChIP assay upon over expression of PRMT6 in K562 cells. PRMT6 occupancy at the CCND1 promoter (transcription start site; TSS) was increased upon over expression of PRMT6. PCR with a primer localized at −4000 served as negative control. G ChIP assay with a LEF1 antibody upon over expression of PRMT6. LEF1 binding to the CCND1 promoter was not altered significantly. H ChIP assay revealed that binding of CTNNB1 (β-Catenin) was reduced upon over expression of PRMT6. Error bars represent the standard deviation from the mean. The P-values were calculated using Student’s t-test from at least three independent measurements. **P < 0.01. I Schematic representation of LEF1/PRMT6 activity on cell cycle genes.