Figure 1 .
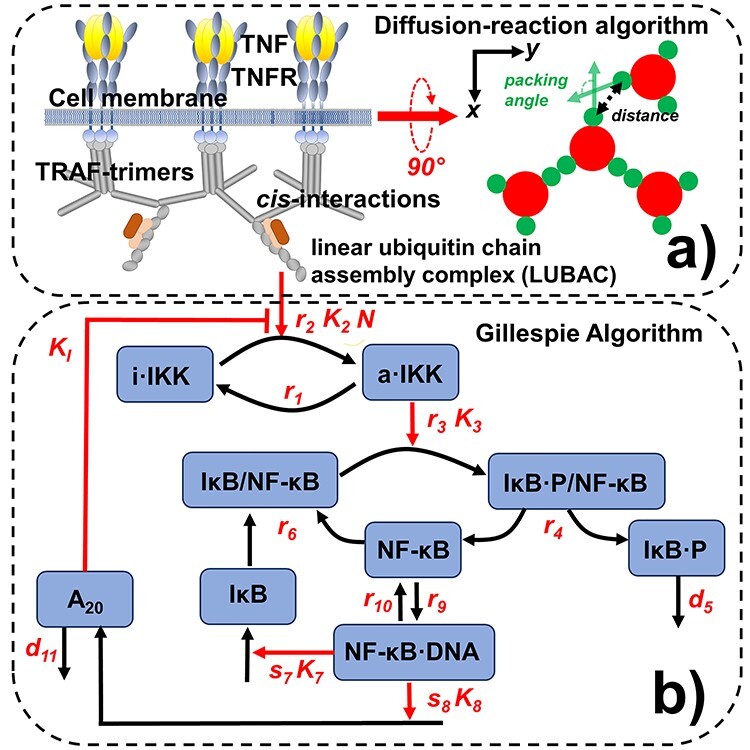
The dynamics of the NF-κB signaling pathway is simulated by a computational method consisting of two coupled systems. The assembly of TRAF-based signaling platform at the membrane proximal region is modeled by a rigid body-based diffusion–reaction algorithm, as shown in (a). We assume that each TRAF trimer has bound to its upstream ligand–receptor complex and therefore is modeled as a single rigid body, in which movements are confined within the 2D membrane proximal area. The formation of a cis-interaction between two TRAF trimers thus facilitates the assembly of linear ubiquitin chain assembly complex (LUBAC). The LUBAC further provide the scaffold to enable the activation of downstream signaling pathway, for which diagram is sketched in (b). The dynamics of the signaling network are simulated by Gillespie algorithm. Finally, the diffusion–reaction and Gillespie algorithms are synchronized under a multiscale simulation framework, as described in the Methods.
