Table 1.
The information about the simulation parameters in the signaling network.
| Index | Description | Mathematical representation | Parameter | Value | Remark |
|---|---|---|---|---|---|
| 1 | Deactivation of IKK |
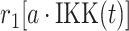
|
r 1 | 0.2 s−1 | Rate of transition |
| 2 | Activation of IKK |
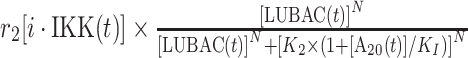
|
r 2 | 0.4 s−1 | Maximal rate of catalysis |
| K 2 | 240 nM | Saturation coefficient of catalysis | |||
| KI | 10 nM | Inhibition coefficient of catalysis | |||
| N | 25 | Hill coefficient | |||
| 3 | Phosphorylation of IκB |
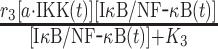
|
r 3 | 0.08 s−1 | Maximal rate of catalysis |
| K 3 | 500 nM | Saturation coefficient of catalysis | |||
| 4 | Dissociation of IκB/NF-κB complex |
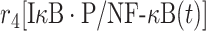
|
r 4 | 0.4 s−1 | Dissociation rate |
| 5 | Degradation of IκB | d 5 | d 5 | 0.8 nMs−1 | Degradation rate |
| 6 | Association of IκB/NF-κB complex |
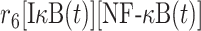
|
r 6 | 0.008 nM−1s−1 | Association rate |
| 7 | Synthesis of IκB |
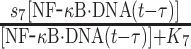
|
s7 | 0.8 s−1 | Maximal rate of synthesis |
| K7 | 50 nM | Saturation coefficient of synthesis | |||
| 8 | Synthesis of A20 |
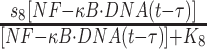
|
s 8 | 0.4 s−1 | Maximal rate of synthesis |
| K 8 | 100 nM | Saturation coefficient of synthesis | |||
| 9 | Association of NF-κB with DNA |
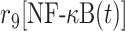
|
r 9 | 0.8 s−1 | Transition rate |
| 10 | Dissociation of NF-κB from DNA |
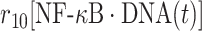
|
r 10 | 0.08 s−1 | Transition rate |
| 11 | Degradation of A20 | d 11 | d 11 | 0.1 nMs−1 | Degradation rate |
