Abstract
Avian leukosis virus subgroup J (ALV-J) has infected a variety of birds, causing major economic losses in China. Understanding the comprehensive criteria of classification and nomenclature of ALV-J would be useful for the investigation of the viral evolution and also for the prevention and control of this infection. An in-depth analysis of the genetic diversity of ALV-J was performed in the present study. Four hundred and seventy-five sequences of the gp85 gene, including thirteen of avian endogenous retrovirus designated ev/J and 462 of ALV-J, were used in the phylogenetic and the evolutionary distance analysis for this classification. The study identified that the current ALV-J strains were divided into two first-order clades (Clades 1 and 2) and three second-order clades (Clades 1.1, 1.2 and 1.3). The current Chinese ALV-J strains are predominantly in Clade 1.3, and the Chinese and Egyptian chicken flocks have been facing the emerging Clade 2 viruses. This system pioneers the classification efforts for ALV-J, which uses Pilot tree for rapid classification of the new isolates and also the addition of possible new clades. The proposed unified classification system will facilitate future studies of ALV-J epidemiology and genetic evolution and of the comparison of sequences obtained across the world.
Keywords: Avian leukosis virus subgroup J, gp85 gene, classification, nomenclature, genetic diversity
1. Introduction
Avian leukosis viruses (ALVs) that infect birds currently comprise eleven envelope subgroups designated A to K (Ma et al. 2020; Wang et al. 2020b). In the last thirty years, the importance of avian leukosis virus subgroup J (ALV-J) infections to avian health has been recognized. ALV-J is an oncogenic exogenous retrovirus and mainly induces neoplastic diseases and immunosuppression in infected birds (Payne, Gillespie, and Howes 1993; Zavala, Cheng, and Jackwood 2007). ALV-J has arisen through multiple recombination events between one or more endogenous and exogenous viruses (Bai, Payne, and Skinner 1995; Benson et al. 1998; Nair and Fadly 2013). ALV-J has the strongest pathogenicity and infectivity among all the subgroups, and it induces the most clinical chicken cases, estimated to account for more than ninety per cent of avian leukosis (AL) (Li et al. 2019). ALV-J was first isolated from commercial meat-type chickens in the United Kingdom in 1988 (Payne, Gillespie, and Howes 1992). Subsequently, ALV-J has caused severe economic loss to the poultry industry worldwide (Payne and Nair 2012). From 1999 to 2001, the virus infected the white-feathered broilers or parent stocks and then spread rapidly throughout China (Cui et al. 2003), and the host range of ALV-J gradually expanding to layer flocks (Cheng et al. 2010; Gao et al. 2012), Yellow-chickens (Sun and Cui 2007; Meng et al. 2018; Li et al. 2019), wild birds (Jiang et al. 2014; Shen et al. 2014; Zeng et al. 2014) and gamecock (Wang et al. 2018b). Generally, commercial chickens mainly include broilers and layers, and the broilers could be classified into White-chickens and Yellow-chickens in China. White-chickens are white-feathered broilers are mainly imported from the world-leading breeding companies from other countries. The Yellow-chickens refer to various indigenous chicken breeds and their hybrid offspring, which normally are colored-feathered chickens (not just yellow-feathered chickens). The broad circulation of ALV-J in poultry has led to significant genetic diversity of the virus and constant emergence of ALV-J. The outbreaks of ALV-J infection are still being reported in China since 2018 (Li et al. 2019; Zhou et al. 2019; Zhang et al. 2020a) even though eradication programs have been established in some big breeding companies (Wei and Cui 2015).
With the molecular methods of viral research, the knowledge of the ALV-J genetic makeup has been improved considerably. ALV belongs to the genus Alpharetrovirus and the family Retroviridae according to the International Committee on Taxonomy of Viruses (ICTV 2020). The proviral DNA arrangement of the genome of ALV-J is 5′ LTR-gag-pol-env-LTR 3′, which consists of approximately 7.2–7.8 kb nucleotides (nt), of which ALV-J env gene is distinct from the other ALV subgroups (Nair and Fadly 2013). The unique pathogenicity of ALV-J is closely related to its env gene, such that the ALV-J env gene could significantly increase the pathogenicity of the recombinant ALV-B (Li et al. 2020a). The envelope protein of ALV-J is encoded by its env gene and can be divided into gp85 and gp37 subunits. The gp85 protein, the viral surface knob-like structure, is closely associated with the process of viral binding and determines the viral envelope subgroup specificity of the ALV (Nair and Fadly 2013; Meng et al. 2016). The gp85 protein is the most variable of the structural proteins and exhibits high genetic diversity in the viral genome (Pan et al. 2012). Recently, it was reported that the bipartite sequence motif in the N (aa 38–131) and C (aa 159–283) termini of gp85 of ALV-J plays a crucial role in receptor binding and viral entry (Zhang et al. 2020b). The gp37 protein is responsible for mediating the fusion of viral protein with the host cell membrane and the C terminus of gp37 plays a vital role in ALV-J pathogenesis (Li et al. 2020b). At present, the need for ALV-J classification has resulted in a variety of methods for isolate differentiation. For example, a phylogenetic analysis based on the gp85 gene sequence found that viruses were related to the genetic background of infected chickens (Li et al. 2018). In the previous research of our research group, isolates from chickens of different genetic backgrounds were sorted into five clusters (Wang et al. 2018b), while the viruses were divided into two different clusters by the full-genome sequences analysis (Wang et al. 2020b). In order to better track the evolutionary history of the ALV-J, we now further propose a unified classification and nomenclature method. When using this approach, the names of the clades of higher order (closer to the tips), bear ancestral information from the lower clades they split from. No report similar to this method has been found. The gp85 gene sequence is usually used for the analysis of molecular epidemiology, and it is also the most detailed gene of ALV-J available in the GenBank database. A previous study showed that the gp85 gene of the env gene could better display the quasispecies diversity of ALV-J than the gp37 gene (Mao et al. 2013). In addition, the isolation and purification of the viruses are time consuming and expensive. The sequences of the gp85 gene of the current ALV-J strains are easier to obtain than their whole genome sequences. Other virus classification systems also use a single gene, such as the HA gene of H5N1 and the F gene of NDV, for classification (WHO/OIE/FAO-H5N1-EWG 2008 2014; Dimitrov et al. 2019). Therefore, it is currently appropriate to use the sequence analysis of the gp85 gene as an index to study the genetic diversity of this virus.
In this study, we employed phylogenetic methods to define the classification of ALV-J based on a total of 475 sequences of the gp85 gene. Our study provides insights into the classification and nomenclature of the current ALV-J strains.
2. Methods
2.1 Viruses sequences used in the study
Nucleotide sequences of the gp85 gene of 475 viruses were collected from our laboratory and from the GenBank National Center for Biotechnology (NCBI, http://www.ncbi.nlm.nih.gov/). One hundred and six sequences were generated (Supplementary Table S1), including eighty-eight sequences from our previous studies sampled during the years 2010–2018 (Lin et al. 2017; Wang et al. 2018a,b; Li et al. 2019; Wang et al. 2020b) and eighteen new sequences sampled during 2017–2020 (GenBank accession numbers: MW491254-MW491271), and 369 sequences with known sampling information, for example dates, locations and host were retrieved from the GenBank database, including 309 ALV-J strains from China sampled during 1999–2020 and forty-seven ALV-J strains from other countries sampled during 1988–2019, plus thirteen avian endogenous retrovirus designated ev/J (Supplementary Table S2). The analysis included nearly the full-length sequences of gp85 (e.g., >900 nt in length) to ensure robust statistical support. An initial alignment of the gp85 gene sequences dataset was aligned using MAFFT v7.149 (Katoh and Standley 2013) and then adjusted manually in BioEdit v7.2.5 (Hall 1999). The final alignment length was 924 nt. Viruses with 100 per cent sequence similarity were identified, giving a final alignment of 406 sequences. Viruses included in Supplementary Tables S1 and S2 that represent other viruses with 100 per cent sequence similarity sequences of the gp85 gene are listed in Supplementary Table S3. In order to further analyze the second-order clades of a dominant clade (Clade 1), a dataset of 387 sequences were further analyzed.
2.2 Phylogenetic analyses
To infer the phylogeny for the first-order clades and the second-order clades of the dominant clades, two phylogenetic trees from two datasets (n = 406 and n = 387) were respectively constructed using IQ-TREE v1.6.12 (Nguyen et al. 2015), and the best-fit model for two datasets were selected by following command: ./iqtree -s infile.phy -m MFP -nt AUTO, and then using the following command: ./iqtree -s infile.phy -m TVM+F+R6 -bb 1000 -nm 5000 -nt AUTO and ./iqtree -s infile.phy -m SYM+R6 -bb 1000 -nm 5000 -nt AUTO, respectively, for the two datasets. Meanwhile, two datasets used a maximum-likelihood (ML) approach in RAxML-HPC v.8 on XSEDE through the CIPRES Science Gateway v.3.3 (Miller, Pfeiffer, and Schwartz 2010) and the following command: raxmlHPC-HYBRID_8.2.12_expanse -f a -n tree -s infile.txt -m GTRGAMMAI -p 12345 -N 1000 -x 12345. All phylogenetic trees were visualized in FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree).
The final Clade 1 and Clade 2 were used to estimate the mean intra-clade distances (nucleotide distances between the first- and second-order clades) and the mean inter-clade distances. The estimates of the average evolutionary distances were inferred using MEGA7 (Kumar, Stecher, and Tamura 2016). The estimates method of average evolutionary distances is similar to the routine described by Dimitrov et al. (2019).
2.3 Pilot tree for rapid preliminary classification
After performing the analyses described above and identifying the main clades, two smaller (pilot) datasets of the gp85 gene sequences (n = 11 and n = 20) were parsed from the complete dataset used in the phylogenetic analyses using IQ-TREE, and the following command: ./iqtree -s infile.phy -m MFP -bb 1000 -nt AUTO. Two pilot datasets were phylogenetically analyzed with the ML method as described above to ensure that the topology inferred using the larger datasets (n = 406 and n = 387) would be maintained if fewer isolates were used.
2.4 Classification and nomenclature criteria of ALV-J
In all analyses, these criteria were used to initially assess the diversity of ALV-J and propose a consensus classification system: an ALV-J classification system that incorporates phylogenetic topology, genetic distances, branch support, and epidemiological independence was used; the names of the clades used Arabic numerals (e.g. 1, 2, 3…); and the ALV-J clades received a numerical-decimal address (Arabic numerals separated by periods) that starts with the Arabic numeral of the branch. Our classification criteria are in alignment with the World Health Organization/World Organization for Animal Health/Food and Agriculture Organization H5N1 Evolution Working Group (WHO/OIE/FAO-H5N1-EWG 2008, 2014) and the NDV Evolution Working Group (Dimitrov et al. 2019). Our classification and nomenclature criteria are summarized in Table 1.
Table 1.
Classification and nomenclature criteria for existing and new viruses.
| No. | Description |
|---|---|
| 1 | A branch, closest to ADOL Line 0 (AF247392), designated as Clade 1.1. Subsequent clades numbered starting from Clade 1.2 (i.e., Clades 1.2–1.9). |
| 2 | Multiple sequence alignment using the MAFFT method. Assignment of different lengths of gp85 gene sequences with the reference virus ADOL Line 0 (AF247392) in batches. |
| 3 | Pilot tree can be used for assigning new viruses to existing clades. |
| 4 | Assignment of viruses into new clades is done based on near-complete gp85 gene from GGAGTTCAT to AGCAGGCGC (or longer) by phylogenetic analysis. |
| 5 | Assignment of viruses into new clades is done by utilizing only a complete dataset of gp85 gene sequences from the GenBank database. |
| 6 | New clades are created based on the phylogenetic tree topology (in a monophyletic groups) using the maximum likelihood method by IQ-TREE and RAxML software. |
| 7 | New clades are created only when four or more independent isolates, without a direct epidemiologic link, are available. |
| 8 | Estimates of evolutionary distances (mean nucleotide distance) between clades are inferred as the number of base substitutions per site from averaging over all sequence pairs between clades using MEGA 7/X software (or a comparable tool) and utilizing the maximum composite likelihood model with rate variation among sites that was modeled with a gamma distribution (shape parameter = 1). |
| 9 | Different first-order clades have an average distance per site above 10% (0.1). Different second-order clades have an average distance per site above 5% (0.05). |
| 10 | The ultrafast bootstrap value at the clade defining node is 70% or above. |
| 11 | All clades are identified using the Arabic numerals system to name. ALV-J clades receive a numerical-decimal address (Arabic numerals separated by periods) that starts with the Arabic numeral of the branch. |
| 12 | If a clade has unresolved topology, low branch support, or insufficient number of isolates, the viruses within this branch are assigned the name of the lower order until all criteria are fulfilled, regardless of the fact that the distance criterion may be correct. |
3. Results
3.1 Phylogenetic analyses
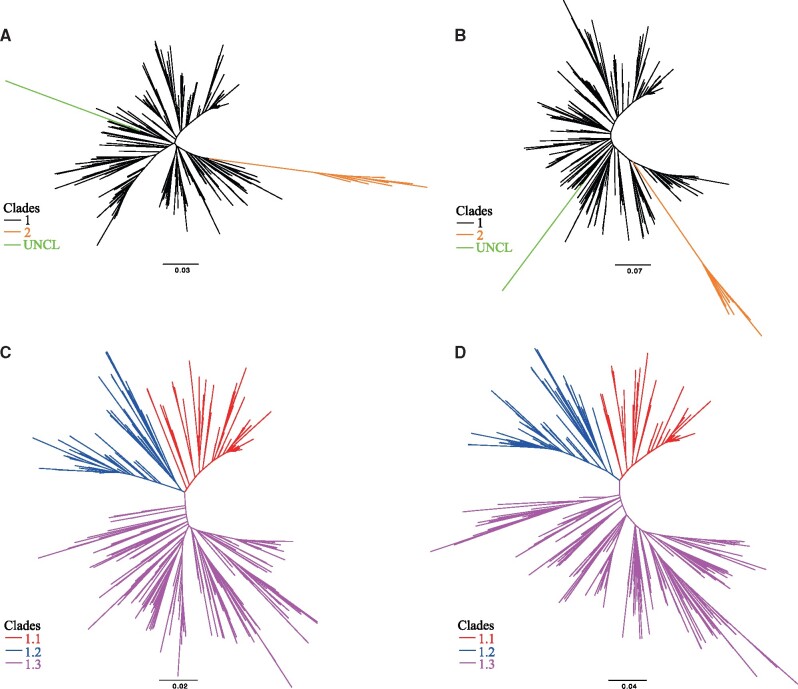
The topologies of the phylogenetic trees, based on the sequences of gp85 gene (Fig. 1A and 1B), showed that all the ALV-J strains were divided into two first-order clades (Clades 1 and 2), respectively. Clade 1 is the main dominant epidemic clade and includes 455 viruses (13 ev/J and 442 ALV-J) from the UK, USA, China, Russia and Pakistan. Clade 2 is increasing and emerging and includes seventeen strains from the USA, China and Egypt (Table 2). In addition, although the genetic distance values of three recombinant strains JL09H01, JL09L01 and JL09DH02 in Jilin, China are >0.1 compared with other clades, they have not been reported in other provinces of China. Thus, no classification has been made (defined as ‘UNCL’) and these strains still need to be continuously monitored. Further phylogenetic analysis was performed to identify the second-order clades of Clade 1. Clade 1 was divided into three clades (Clades 1.1, 1.2 and 1.3) (Fig. 1C and 1D, Supplementary Tables S1 and S2). Clade 1.1, which has the widest range of sources, includes forty-four (44/156) strains from White-chickens, nineteen strains from layers, fifty-six strains from Yellow-chickens, one strain from the contaminated live vaccine, two strains from wild birds, five strains from gamecocks and plus thirteen ev/J, etc. Clade 1.2 primarily includes forty (40/88) strains from layers, twenty-eight strains from Yellow-chickens and twelve strains from wild birds. Clade 1.3 mainly includes forty-one (41/211) strains from White-chickens, twenty strains from layers and one hundred and twenty-one strains from Yellow-chickens. The current Chinese dominant ALV-J strains are in Clade 1.3. The strains from our laboratory were classified as clades 1.1 (14/106), 1.2 (14/106) and 1.3 (78/106).
Figure 1.
Phylogenetic analysis was performed based on the nucleotide sequences of the gp85 gene of ALV-J and ev/J. (A and B) Two radial phylogenetic trees were constructed based on the gp85 gene sequences (n = 406) of viruses representing ALV-J and ev/J to identify the first-order clades with IQ-TREE and RAxML software, respectively. (C and D) Two radial phylogenetic trees were constructed based on the gp85 gene sequences (n = 387) of viruses representing ALV-J and ev/J to identify the second-order clades of Clade 1 with IQ-TREE and RAxML software, respectively. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site.
Table 2.
Details of strains obtained from Clade 2.
| No. | Strains | Accession no. | Host | Location | Time |
|---|---|---|---|---|---|
| 1 | ADOL-7501 | AY027920 | W | USA | 1997 |
| 2 | 1696 | AF247384 | W | USA | 1997 |
| 3 | 6803 | AF247388 | W | USA | 1997 |
| 4 | 6827 | AF247389 | W | USA | 1997 |
| 5 | AF88 | AF247390 | W | USA | 1997 |
| 6 | UD-J1 | AF305091 | N | USA | 2000 |
| 7 | SCSM00 | KF796652 | Y | Sichuan, China | 2011-10 |
| 8 | GDQJ-1 | KU254611 | N | Guangdong, China | 2014 |
| 9 | HB18XH01 | MN735298 | Y | Hebei, China | 2018 |
| 10 | SH18JY01 | MN735306 | Y | Shanghai, China | 2018 |
| 11 | SH18JY02 | MN735307 | Y | Shanghai, China | 2018 |
| 12 | QL1 | MN496121 | L | Egypt | 2019-02 |
| 13 | QL2 | MN496122 | L | Egypt | 2018-11 |
| 14 | QL3 | MN496123 | L | Egypt | 2018-04 |
| 15 | QL4 | MN496124 | L | Egypt | 2019-05 |
| 16 | QL5 | MN496125 | L | Egypt | 2019-12 |
| 17 | QL6 | MN496126 | L | Egypt | 2018-08 |
W: White-chickens; L: layers; Y: Yellow-chickens; N: not available.
3.2 Evolutionary distances inferences
Distance analysis (Supplementary Tables S4 and S5) shows that the genetic distance between the two first-order clades was greater than ten per cent, and that among the three second-order clades was greater than five per cent. Within the second-order clade of Clade 1, the gp85 genes of Clade 1.1 are conservative (<4% variation) when compared with those of Clades 1.2 and 1.3. The main reason for this is that Clade 1.1 is in a cluster with the conservative ev/J. The estimations of evolutionary distances also confirmed that the classification of phylogenetic trees is appropriate.
3.3 Pilot tree and dataset
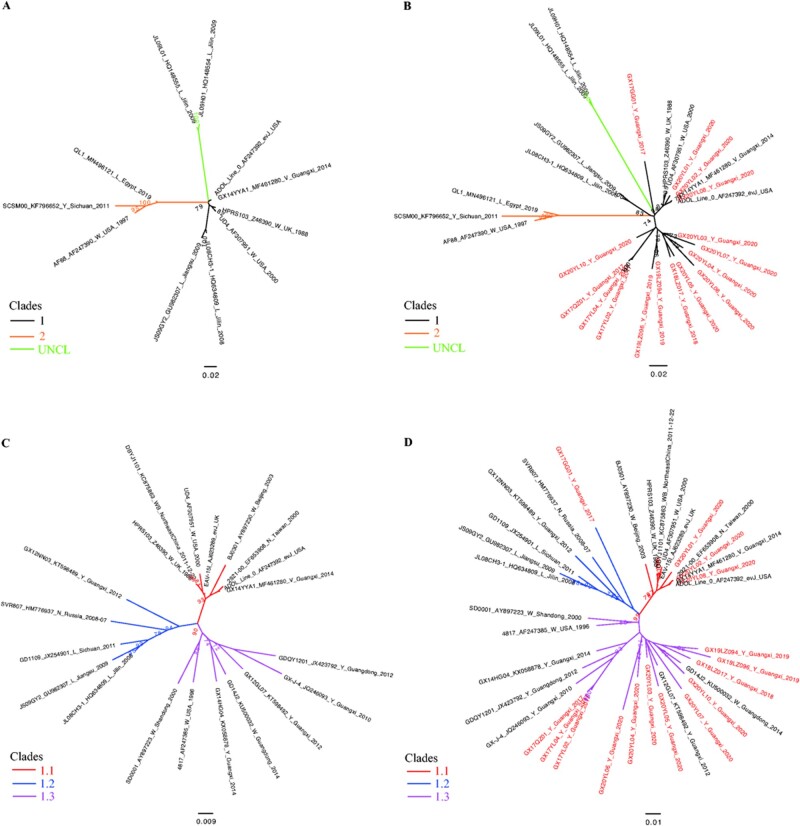
Here, we needed to find a rapid, preliminary method for clade identification of the new isolates, utilizing a phylogenetic tree with a lesser number of sequences that still provides topology consistent to the larger tree and clearly decreases the time for the tree construction. The eleven and twenty representative sequences of viruses were respectively used to perform ML phylogenetic reconstruction to distinguish the first-order clades and the second-order clades of Clade 1 (Fig. 2 and Table 3). These generated trees were consistent with the ML trees inferred using the larger gp85 gene dataset in Fig. 1. Thus, this pilot dataset can be utilized for rapid preliminary classification of the new isolates.
Figure 2.
Classification “pilot” tree. (A and C) Phylogenetic analysis are based on the gp85 genes of the selected viruses representing the first-order clade (n = 11) and the second-order clade (n = 20), respectively. (B and D) Use the newly isolated strains during 2017–2020 to test the topological structure of the tree. New isolates assigned in this current study are highlighted in red font. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site.
Table 3.
Details of the representative viruses in the pilot tree of clades.
| Number of viruses | Clades | Representative viruses (Accession no.) |
|---|---|---|
| 11 | 1 | ADOL Line 0 (AF247392), HPRS103 (Z46390), GX14YYA1 (MF461280), UD4 (AF307951), JL08CH3-1 (HQ634809), JS09GY2 (GU982307) |
| 2 | AF88 (AF247390), QL1 (MN496121), SCSM00 (KF796652) | |
| UNCL | JL09H01 (HQ148554), JL09L01 (HQ148555) | |
| 20 | 1.1 | ADOL Line 0 (AF247392), HPRS103 (Z46390), EAV-15I (AJ623289), 2921/00 (EF653908), DBYJ1101 (KC875863), GX14YYA1 (MF461280), UD4 (AF307951), BJ0301 (AY897230) |
| 1.2 | JL08CH3-1 (HQ634809), JS09GY2 (GU982307), SVR807 (HM776937), GD1109 (JX254901), GX12NN03 (KT598489) | |
| 1.3 | GD14J2 (KU500032), GX12GL07 (KT598492), GDQY1201 (JX423792), 4817 (AF247385), GX14HG04 (KX058878), GX-J-4 (JQ246093), SD0001 (AY897223) |
3.4 Nomenclature criteria
To ensure consistent naming, nomenclature criteria guidelines for naming clades are also outlined in Table 1. Most importantly, when using this approach, the names of the clades of higher order (closer to the tips) bear ancestral information from the lower clades they split off from, which helps to understand the epidemiology of ALV-J.
4. Discussion
ALV-J-related studies have been ongoing for more than thirty years; however, understanding the criteria for the classification and nomenclature of the viruses has remained poorly investigated. Although progress has been made in the control by the ALV eradication programs implemented since 2008 in China (Wei and Cui 2015), it still remains a threat, with quite a few cases in Chinese indigenous chickens reported every year. This comprehensive analysis of 475 sequences of the gp85 gene over the past three more decades will facilitate the understanding of the evolution of the virus, and the subsequent disease prevention and control.
In order to better track the origin of the virus, a more detailed classification and nomenclature method has been established here. Similar to our previous findings (Wang et al. 2018b, 2020b), viruses form independent clusters that are related to correspondingly host genetic backgrounds. In this study, we have improved the accuracy of the results in terms of classification methods and can now trace the source of the virus in terms of this nomenclature method. We first used the MAFFT software for sequence alignment, which aligned the different lengths of gp85 gene sequences with reference virus ADOL Line 0 (AF247392) in batches, removed the identical sequence using IQ-TREE and BioEdit software and then adopted the ML phylogenies generated by IQ-TREE and RAxML software, thereby greatly improving the stability of the evolutionary tree topologies. Additionally, some limitations of the methods are considered when interpreting our findings, but only 4.4 per cent (17/387) of all strains are inconsistent in classification by using the two methods. This analysis of the phylogeny did not change our conclusion. A recent study revealed that 18.1 per cent (3515/19,414) gene alignments from fifteen phylogenomic datasets yielded irreproducible phylogenies between two replicates using IQ-TREE, while 9.3 per cent (1813) genes yielded irreproducible phylogenies between two replicates using RAxML-NG (Shen et al. 2020). Compared with the results of IQ-TREE, the results of RAxML are more stable. Thus, the classification in this study mainly refers to the results of the RAxML. Selecting high-quality nucleotide sequences and sequence alignment using the MAFFT software and the ML phylogenies generated using multiple software will facilitate constructing a topologically stable evolutionary tree. The clades of the nomenclature suggested here are divided into three levels at most, including the first-order clades, the second-order clades and the third-order clades, so that it is easy to remember. ALV-J clades receive a numerical-decimal address (Arabic numerals separated by periods) that starts with the Arabic numeral of the branch.
In this study, the currently available ALV-J strains could be divided into two first-order clades (Clades 1 and 2) and three second-order clades (Clades 1.1, 1.2, 1.3) (Fig. 1). The evolutionary tree of Clade 1 made by our classification shows the spread of ALV-J from White-chickens to layers and Yellow-chickens. Clade 1.1 is a conservative cluster (Supplementary Tables S4 and S5). Clade 1.1 includes not only ALV-J isolates from various sources but also the 13 ev/J. Previous studies showed ALV-J is a multiple recombinant of the env sequence of ev/J and the exogenous avian leukosis virus (Bai, Payne, and Skinner 1995; Benson et al. 1998; Nair and Fadly 2013). Some of the ALV-J viruses in this cluster are closely related with the ev/J may be the relatively primitive viruses that have just been recombined the env sequence from ev/J. Also, it is worth noting that our isolate GX14YYA1 originated from the contaminated live commercial vaccine (Wang et al. 2018a) is in this cluster. Some of these strains are highly similar to the ALV-contaminated live vaccines isolate, and it is possible that they were transmitted through the contamination live vaccines and is contributing to the inability to eradicate ALV. They may contaminate the live vaccines produced by using the chicken embryo and/its cells. Clade 1.2 is mainly a branch of strains originating in layers. Clade 1.3 is dominated by the Chinese field strains originating in Yellow-chickens. Thirteen isolates from our laboratory were isolated by detecting the P27 antigen of ALV on the DF-1 cell cultures inoculated with the clinical tumor tissue samples at diagnosis and the sterile plasma samples of the birds at eradication testing during the years 2018–2020 (Supplementary Table S1). These were gathered in Clades 1.1 and 1.3, respectively, but the major clinical sample isolates (3/5) and the plasma sample isolates (7/8) were in Clade 1.3, revealing that incomplete eradication of ALV is one of the main reasons for the clinical AL cases. In addition, the ALV-J from the outbreaks in 2018–2019 in China had the closest genetic relationship with the isolate GD14J2 isolated from the White-chickens that have the origins from other countries (Ma et al. 2019; Zhang et al. 2020a). The Sichuan, Guangdong, Hebei and Shanghai isolates in Clade 2 obtained during the years 2011–2018 were found to be emerging, and which was most likely spread to China by the introduction of the White-chickens breeders from abroad (Wang et al. 2020a; Yu et al. 2014). Previously, few researchers paid attention to the pathogenicity and transmissibility of the six American strains (ADOL-7501, 1696, 6803, 6827, AF88 and UD-J1) in Clade 2. Now Clade 2 strains have been found in China and Egypt, where Clade 2 are gradually increasing and emerging, so it is necessary to continually monitor them (Table 2). Hence, four main clades (1.1, 1.2, 1.3 and 2) of ALV-J strains were revealed in China. There are three main causes for the increased genetic diversity of isolates from the commercial chicken flocks: the use of ALV-contaminated live vaccine/vaccines; the introduction of breeder chickens from abroad; and the incomplete eradication in the breeder flocks.
In conclusion, we first proposed the definition of the current ALV-J strains based on the gp85 gene that were divided into two first-order clades and three second-order clades. The current Chinese dominant ALV-J strains belong to Clade 1.3 and the Chinese and Egyptian chicken flocks have been facing the emerging Clade 2 viruses. This system includes comprehensive criteria for the classification of new ALV-J isolates and also the addition of the possible new clades. Our classification can play an irreplaceable role in tracking the evolution of viruses and in monitoring the spread of viruses currently.
Supplementary data
Supplementary data are available at Virus Evolution online.
Supplementary Material
Acknowledgements
This work was supported by the Guangxi Special Funding on Science and Technology Research [AA17204057], the Guangxi Program for Modern Agricultural Industry Technical System Construction-Chicken Industry [nycytxgxcxtd-19-03], the Shandong Provincial Natural Science Foundation [ZR2019BC047]. The manuscript was kindly reviewed by Dr. Richard Roberts, Aurora, CO 80014, USA.
Data availability
The data generated or analyzed during this study are available within the article and its supporting information.
Author contributions
Qiaomu Deng completed the draft; Qiaomu Deng, Min Li and Peikun Wang analyzed the data; Ping Wei provided the funding of research, reviewed and approved the final manuscript; all authors participated in this study.
Ethics statement
This study was approved by the Animal Welfare and the Animal Experimental Ethical Committee of Guangxi University, and conventional animal welfare regulations and standards were taken into account. The study, using a modern molecular approach, did not involve animal and/or human work.
Conflict of Interest: None declared.
References
- Bai J., Payne L. N., Skinner M. A. (1995) ‘HPRS-103 (Exogenous Avian Leukosis Virus, Subgroup J) Has an Env Gene Related to Those of Endogenous Elements EAV-0 and E51 and an E Element Found Previously Only in Sarcoma Viruses’, Journal of Virology, 69: 779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S. J. et al. (1998) ‘The Unique Envelope Gene of the Subgroup J Avian Leukosis Virus Derives from ev/J Proviruses, a Novel Family of Avian Endogenous Viruses’, Journal of Virology, 72: 10157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. et al. (2010) ‘Tumors Associated with Avian Leukosis Virus Subgroup J in Layer Hens during 2007 to 2009 in China’, The Journal of Veterinary Medical Science, 72: 1027–33. [DOI] [PubMed] [Google Scholar]
- Cui Z. et al. (2003) ‘Comparison of Chinese Field Strains of Avian Leukosis Subgroup J Viruses with Prototype Strain HPRS-103 and United States Strains’, Avian Diseases, 47: 1321–30. [DOI] [PubMed] [Google Scholar]
- Dimitrov K. M. et al. (2019) ‘Updated Unified Phylogenetic Classification System and Revised Nomenclature for Newcastle Disease Virus’, Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 74: 103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. et al. (2012) ‘Molecular Epidemiology of Avian Leukosis Virus Subgroup J in Layer Flocks in China’, Journal of Clinical Microbiology, 50: 953–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999) ‘BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT’, Nucleic Acids Symposium Series, 41: 95–8. [Google Scholar]
- ICTV. (2020) Virus Taxonomy: 2019 Release. International committee on taxonomy of viruses. <https://talk.ictvonline.org/taxonomy/> accessed December 2020.
- Jiang L. et al. (2014) ‘Genetic Diversity and Phylogenetic Analysis of Glycoprotein gp85 of Avian Leukosis Virus Subgroup J Wild-Bird Isolates from Northeast China’, Archives of Virology, 159: 1821–6. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016) ‘MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets', Molecular ’, Molecular Biology and Evolution, 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2019) ‘The Emergence of the Infection of Subgroup J Avian Leucosis Virus Escalated the Tumour Incidence in Commercial Yellow Chickens in Southern China in Recent Years’, Transboundary and Emerging Diseases, 66: 312–6. [DOI] [PubMed] [Google Scholar]
- Li J. et al. (2018) ‘Characterization of Avian Leukosis Virus Subgroup J Isolated between 1999 and 2013 in China’, Poultry Science, 97: 3532–9. [DOI] [PubMed] [Google Scholar]
- Li Q. et al. (2020a) ‘Recombinant Subgroup B Avian Leukosis Virus Combined with the Subgroup J Env Gene Significantly Increases Its Pathogenicity’, Veterinary Microbiology, 250: 108862. [DOI] [PubMed] [Google Scholar]
- Li T. et al. (2020b) ‘Gp37 Regulates the Pathogenesis of Avian Leukosis Virus Subgroup J via Its C Terminus’, Journal of Virology, 94: e02180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. et al. (2017) ‘Full-Length Genome Sequence Analysis of Four Subgroup J Avian Leukosis Virus Strains Isolated from Chickens with Clinical Hemangioma’, Virus Genes, 53: 868–75. [DOI] [PubMed] [Google Scholar]
- Ma M. et al. (2020) ‘Molecular Characterization of Avian Leukosis Virus Subgroup J in Chinese Local Chickens between 2013 and 2018’, Poultry Science, 99: 5286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M. G. et al. (2019) ‘Full-Length Genome Sequence Analysis of Subgroup J Avian Leukosis Viruses Isolated from Imported White Feather Broiler Breeders’, Chinese Journal of Preventive Veterinary Medicine, 41: 750–4 (in Chinese). [Google Scholar]
- Mao Y. et al. (2013) ‘Different Quasispecies with Great Mutations Hide in the Same Subgroup J Field Strain of Avian Leukosis Virus’, Science China Life Sciences, 56: 414–20. [DOI] [PubMed] [Google Scholar]
- Meng F. et al. (2016) ‘A Deep Sequencing Reveals Significant Diversity among Dominant Variants and Evolutionary Dynamics of Avian Leukosis Viruses in Two Infectious Ecosystems’, BMC Veterinary Research, 12: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F. et al. (2018) ‘Characterization of Subgroup J Avian Leukosis Virus Isolated from Chinese Indigenous Chickens’, Virology Journal, 15: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010) ‘Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees’, Gateway Computing Environments Workshop (GCE), 14: 1–8. [Google Scholar]
- Nair V., Fadly A. M., (2013) ‘Leukosis/Sarcoma Group’, in Swayne D.E.et al. (eds.) Diseases of Poultry, 13th edn., pp. 553–92. Ames, IA: Wiley, Blackwell. [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. et al. (2012) ‘Genetic Diversity and Phylogenetic Analysis of Glycoprotein GP85 of ALV-J Isolates from Mainland China between 1999 and 2010: Coexistence of Two Extremely Different Subgroups in Layers’, Veterinary Microbiology, 156: 205–12. [DOI] [PubMed] [Google Scholar]
- Payne L. N., Gillespie A. M., Howes K. (1992) ‘Myeloid Leukaemogenicity and Transmission of the HPRS-103 Strain of Avian Leukosis Virus’, Leukemia, 6: 1167–76. [PubMed] [Google Scholar]
- Payne L. N., Gillespie A. M., Howes K. (1993) ‘Recovery of Acutely Transforming Viruses from Myeloid Leukosis Induced by the HPRS-103 Strain of Avian Leukosis Virus’, Avian Diseases, 37: 438–50. [PubMed] [Google Scholar]
- Payne L. N., Nair V. (2012) ‘The Long View: 40 Years of Avian Leukosis Research’, Avian Pathology: Journal of the W.V.P.A, 41: 11–9. [DOI] [PubMed] [Google Scholar]
- Shen X. X. et al. (2020) ‘An Investigation of Irreproducibility in Maximum Likelihood Phylogenetic Inference’, Nature Communications, 11: 6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. et al. (2014) ‘Genetic Mutations of Avian Leukosis Virus Subgroup J Strains Extended Their Host Range’, Journal of General Virology, 95: 691–9. [DOI] [PubMed] [Google Scholar]
- Sun S., Cui Z. (2007) ‘Epidemiological and Pathological Studies of Subgroup J Avian Leukosis Virus Infections in Chinese Local "Yellow" Chickens’, Avian Pathology : Journal of the W.V.P.A, 36: 221–6. [DOI] [PubMed] [Google Scholar]
- Wang G. Y. et al. (2020a) ‘Isolation, Identification and Evolution Analysis of Subgroup J Avian Leukosis Virus Isolated from Guangdong Local Breed Chicken’, China Poultry, 42: 28–33 (in Chinese). [Google Scholar]
- Wang P. et al. (2020b) ‘Full-Length cDNA Sequence Analysis of 85 Avian Leukosis Virus Subgroup J Strains Isolated from Chickens in China during the Years 1988-2018: Coexistence of 2 Extremely Different Clusters That Are Highly Dependent upon Either the Host Genetic Background or the Geographic Location’, Poultry Science, 99: 3469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. et al. (2018a) ‘Full-Length Genome Sequence Analysis of an Avian Leukosis Virus Subgroup J (ALV-J) as Contaminant in Live Poultry Vaccine: The Commercial Live Vaccines Might Be a Potential Route for ALV-J Transmission’, Transboundary and Emerging Diseases, 65: 1103–6. [DOI] [PubMed] [Google Scholar]
- Wang P. et al. (2018b) ‘Diversity and Evolution Analysis of Glycoprotein GP85 from Avian Leukosis Virus Subgroup J Isolates from Chickens of Different Genetic Backgrounds during 1989-2016: Coexistence of Five Extremely Different Clusters’, Archives of Virology, 163: 377–89. [DOI] [PubMed] [Google Scholar]
- Wei P., Cui Z. Z. (2015) ‘The Harm of Avian Leukemia Virus and Salmonella pullorum to Local Chickens and the Measures of the Eradication’, China Poultry, 37: 1–4 (in Chinese). [Google Scholar]
- WHO/OIE/FAO-H5N1-EWG. (2008) ‘Toward a Unified Nomenclature System for Highly Pathogenic Avian Influenza Virus (H5N1)’, Emerging Infectious Diseases, 14: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO/OIE/FAO-H5N1-EWG. (2014) ‘Revised and Updated Nomenclature for Highly Pathogenic Avian Influenza A (H5N1) Viruses’, Influenza and Other Respiratory Viruses, 8: 384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. H. et al. (2014) ‘Molecular Epidemiology of Subgroup J Avian Leukosis Virus from Sichuan Area during 2011 to 2012’, Chinese Journal of Preventive Veterinary Medicine, 36: 404–6 (in Chinese). [Google Scholar]
- Zavala G., Cheng S., Jackwood M. W. (2007) ‘Molecular Epidemiology of Avian Leukosis Virus Subgroup J and Evolutionary History of Its 3' Untranslated Region’, Avian Diseases, 51: 942–53. [DOI] [PubMed] [Google Scholar]
- Zeng X. et al. (2014) ‘Molecular Characteristics of the Complete Genome of a J-Subgroup Avian Leukosis Virus Strain Isolated from Eurasian Teal in China’, Virus Genes, 49: 250–8. [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. (2020a) ‘Molecular Characteristics of the Re-Emerged Avian Leukosis Virus in China, 2018-2019’, Transboundary and Emerging Diseases, 67: 1141–51. [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. (2020b) ‘The Bipartite Sequence Motif in the N and C Termini of gp85 of Subgroup J Avian Leukosis Virus Plays a Crucial Role in Receptor Binding and Viral Entry’, Journal of Virology, 94: e01232-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D. et al. (2019) ‘Outbreak of Myelocytomatosis Caused by Mutational Avian Leukosis Virus Subgroup J in China, 2018’, Transboundary and Emerging Diseases, 66: 622–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated or analyzed during this study are available within the article and its supporting information.