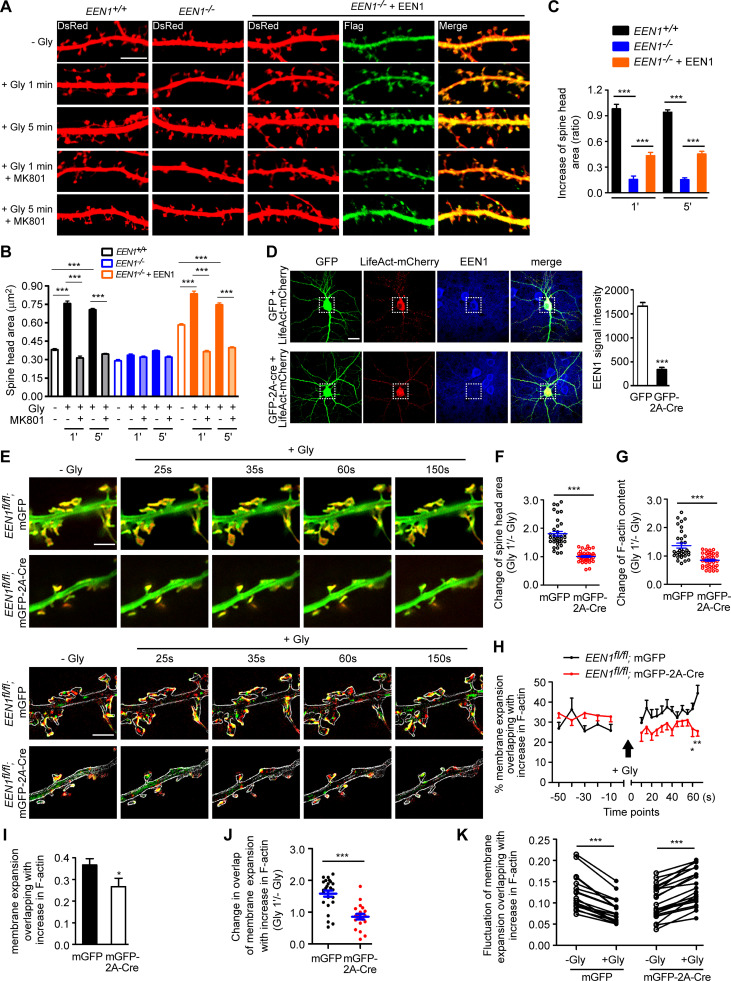
Figure 1.
Ablation of endophilin A1 abolishes spine expansion and actin polymerization in the acute phase of sLTP. (A) Rescue of sLTP in EEN1−/− neurons. Mouse hippocampal neurons cotransfected with pLL3.7-DsRed (volume marker) and pCMV-Tag2B (FLAG vector) or pCMV-Tag2B-endophilin A1 (FLAG-EEN1) were pretreated with DMSO (vehicle control) or MK801 (NMDAR antagonist), and cLTP was performed using glycine (Gly) on DIV16. Neurons were fixed 1 min or 5 min after glycine application, immunostained for FLAG, and imaged by confocal microscopy. Scale bar, 5 µm. (B and C) Quantification of spine size (B) and changes of spine size (C) in A. Data are expressed as mean ± SEM for each group (EEN1+/+: n [cells] = 20, n [spines] = 851 for − Gly; n = 15, n = 571 for + Gly 1 min; n = 20, n = 871 for + Gly 5 min; n = 14, n = 568 for + Gly 1 min + MK801; n = 17, n = 668 for + Gly 5 min + MK801. EEN1−/−: n = 20, n = 831 for − Gly; n = 13, n = 563 for + Gly 1 min; n = 16, n = 614 for + Gly 5 min; n = 15, n = 573 for + Gly 1 min + MK801; n = 15, n = 569 for + Gly 5 min + MK801. EEN1−/− + EEN1: n = 19, n = 731 for − Gly; n = 14, n = 581 for + Gly 1 min; n = 16, n = 654 for + Gly 5 min; n = 14, n = 572 for + Gly 1 min + MK801; n = 15, n = 604 for + Gly 5 min + MK801). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001. (D) Cre-mediated KO in EEN1fl/fl hippocampal neurons. Neurons from an EEN1fl/fl mouse were transfected with constructs expressing LifeAct-mCherry and GFP or both GFP and the Cre recombinase (GFP-2A-Cre) on DIV12 and immunostained for EEN1 on DIV16. Scale bar, 20 µm. Quantitative data were expressed as mean ± SEM for each group (n = 15 for GFP, n = 13 for GFP-2A-Cre). P values were calculated using two-tailed unpaired t test. ***, P < 0.0001. (E) GI-SIM live imaging of EEN1fl/fl hippocampal neurons expressing LifeAct-mCherry and mGFP or mGFP-2A-Cre. Glycine was applied after imaging for 1 min, and imaging was continued for 5 more minutes. Upper panels are representative still images before and after glycine application. Lower panels are color-coded images showing regions of membrane expansion (increase in mGFP signals, green) and actin polymerization (increase in F-actin signals, red) in dendritic spines (outlined with the Find Edges tool of ImageJ). Scale bar, 2 µm. (F and G) Quantitative analysis of spine size increase (F) or F-actin enrichment in spines (G) of EEN1fl/fl;mGFP (WT) and EEN1fl/fl;mGFP-2A-Cre (KO) neurons imaged by GI-SIM. Data are plotted along with the mean ± SEM for each group (n = 5, n = 36 for EEN1fl/fl;mGFP, n = 5, n = 42 for EEN1fl/fl;mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. ***, P < 0.0001. (H) Quantification of fractions of expanded membrane overlapping with actin polymerization in individual spines before and after glycine application at 10-s (before) or 5-s (after) intervals. Data for each point are expressed as mean ± SEM (n = 5, n = 18 for EEN1fl/fl;mGFP, n = 5, n = 20 for EEN1fl/fl;mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. *, P < 0.05; **, P < 0.01. (I) Quantification of fractions of membrane expansion overlapping with actin polymerization at 1 min after glycine application. Data are expressed as mean ± SEM (n = 5, n = 24 for mGFP; n = 5, n = 20 for mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. *, P < 0.05. (J) Changes in the extent of overlap between membrane expansion and actin polymerization 1 min after glycine application in individual spines. Data are normalized to the time point right before glycine application. The data are plotted along with the mean ± SEM for each group (n = 5, n = 24 for mGFP; n = 5, n = 20 for mGFP-2A-Cre). P values were calculated using two-tailed unpaired t test. ***, P < 0.0001. (K) Mean fluctuation of the overlap between membrane expansion and actin polymerization in individual spines within 1 min before and after glycine application. n = 5, n = 18 for mGFP; n = 5, n = 20 for mGFP-2A-Cre. P values were calculated using two-tailed paired t test. ***, P < 0.0001.