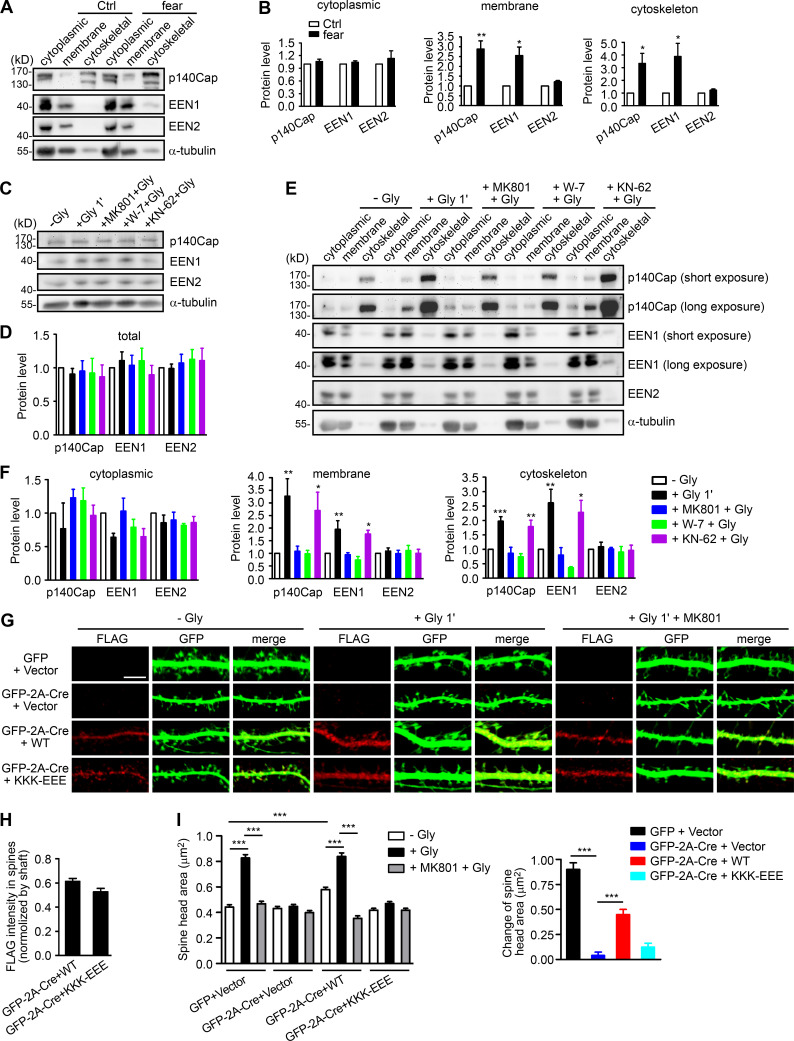
Figure 5.
Ca2+/calmodulin enhances the association of endophilin A1 and p140Cap with both membrane and cytoskeleton upon LTP induction. (A) Immunoblotting of subcellular fractionations of mouse hippocampi from naive or fear conditioned animals. (B) Quantification of protein levels of p140Cap, EEN1, or A2 in cytoplasmic, membrane, and cytoskeleton fractions in A. n = 6 animals for each group. P values were calculated using two-tailed paired t test. *, P < 0.05; **, P < 0.01 when compared with Ctrl. (C) DIV16 neurons were pretreated with DMSO, MK801, W-7, or KN-62 and induced cLTP with glycine for 1 min, lysed, and subjected to SDS-PAGE and immunoblotting. (D) Quantification of total protein levels in C. n = 5 independent experiments. (E) Effects of MK801, W-7, and KN-62 on subcellular distribution of proteins. DIV16 neurons were pretreated with DMSO, MK801, W-7, or KN-62 and induced cLTP with glycine for 1 min, collected, and subjected to subcellular fractionation. (F) Quantification of protein levels in subcellular fractions in E. n = 5 independent experiments. P values were calculated using one-way ANOVA by Dunnett post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 when compared with − Gly. (G) EEN1fl/fl hippocampal neurons cotransfected with GFP or GFP-2A-Cre and FLAG vector or GFP-2A-Cre and FLAG-EEN1 WT or KKK-EEE expression constructs on DIV12 were pretreated with DMSO or MK801 and induced cLTP on DIV16 with glycine for 1 min, immunostained for FLAG, and imaged by confocal microscopy. Scale bar, 5 µm. (H) Spine/shaft distribution of EEN1 KKK-EEE compared with WT. Data are expressed as mean ± SEM for each group (n = 10, n = 202 for Cre + WT; n = 11, n = 216 for Cre + KKK-EEE). P values were calculated using two-tailed unpaired t test. (I) Quantification of spine size and changes in spine size in G. Data are expressed as mean ± SEM for each group (GFP + vector: n = 11, n = 205 for − Gly; n = 11, n = 213 for + Gly; n = 11, n = 227 for + MK801 + Gly; Cre + vector: n = 11, n = 197 for − Gly; n = 11, n = 194 for + Gly; n = 11, n = 195 for + MK801 + Gly; Cre + WT: n = 10, n = 202 for − Gly; n = 11, n = 226 for + Gly; n = 10, n = 193 for + MK801 + Gly; Cre + KKK-EEE: n = 11, n = 216 for − Gly; n = 11, n = 211 for + Gly; n = 10, n = 190 for + MK801 + Gly). P values were calculated using one-way ANOVA by Newman–Keuls post hoc test. ***, P < 0.001.