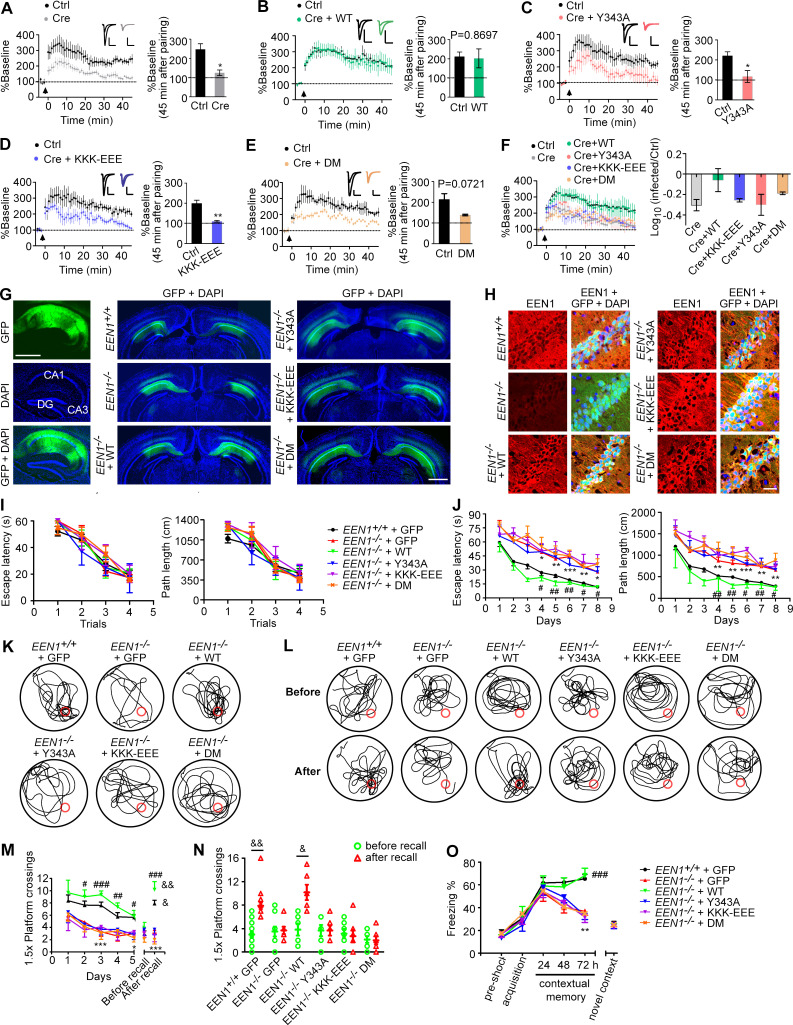
Figure 7.
The calmodulin-, membrane-, and p140Cap-binding capacities of endophilin A1 are required for LTP and long-term memory. (A–F) Rescue of the LTP impairment phenotype in EEN1 KO neurons by EEN1 WT and mutants. AAVs expressing Cre (AAV-mCherry-2A-Cre) and GFP (AAV-EGFP) or Cre, GFP, and EEN1 WT or mutant (AAV-EGFP-2A-EEN1) were stereotaxically injected into the CA1 regions of EEN1fl/fl mouse brain at P0. Acute hippocampal slices were prepared on P14–P21 for dual recording analysis of LTP. Shown are pairwise comparisons of LTP in noninfected (Ctrl) and infected neurons of the same slice. For Cre-expressing neurons (Cre) versus Ctrl (A), six recording pairs from three mice, marked as n = 6/3, were analyzed. n = 10/5 (B), n = 5/4 (C), n = 5/3 (D), and n = 4/3 (E). Bar graphs show percentage of baseline at 45 min after pairing. A summary of rescue effects of EEN1 WT and mutants is shown in F. Data are expressed as mean ± SEM for each group. P values were calculated using two-tailed paired t test. *, P < 0.05; **, P < 0.01. (G) AAV was stereotaxically injected into the brain CA1 regions of EEN1+/+ to express GFP alone, EEN1−/− to express GFP alone, or GFP and EEN1 WT, KKK-EEE, Y343A, or DM. Left panels are images of hippocampal sagittal section with GFP signals and DAPI labeling of nuclei. Center and right panels are coronal brain sections showing bilateral injection of AAV. Scale bar, 1 mm. (H) Immunofluorescence staining of EEN1 in CA1 neurons of brain slices from mice in G. Scale bar, 50 µm. (I–N) Morris water maze test of AAV-injected mice. Shown are escape latency or distance traveled before escaping to the platform in the visible platform (I) and invisible platform (J) training, the swim trace in probe trial 3 and recall following training once again 1 mo after training (K and L), number of crossings within the 1.5× platform area in 5-d probe trial and recall test (M), and number of crossings within the 1.5× platform area before and after recall (N). (O) Freezing behavior in AAV-injected mice subjected to contextual fear conditioning. All error bars represent the SEM in I, J, and M–O. Number of animals: 15 EEN1+/+ + GFP, 10 EEN1−/− + GFP, 6 EEN1−/− + EEN1, 6 EEN1−/− + Y343A, 7 EEN1−/− + KKK-EEE, 7 EEN1−/− + DM. P values were calculated using one-way ANOVA by Newman–Keuls post hoc test or two-tailed paired t test. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 when compared with EEN1+/+ + GFP; #, P < 0.05; ##, P < 0.01; and ###, P < 0.001 when compared with EEN1−/− + GFP; and &, P < 0.05 and &&, P < 0.01 when compared with before recall (two-tailed paired t test).