Abstract
Intensive, prolonged exercise is known to induce gastrointestinal disorders such as diarrhea, with gut dysbiosis suggested as being one of the causatives. In the present study, we wanted to investigate the relationship between intensive exercise and the gut microbiota status. To that end, the microbiota, the moisture content and the bacterial metabolites (e.g., organic acids) of female endurance runners (n = 15) and those of non-athletic but healthy, age-matching female controls (n = 14) were compared. The analysis of the gut microbiota analysis showed that, unlike control subjects, female endurance runners had distinct microbiotas, with some bacteria found in higher abundances likely being involved in gut inflammation. The concentration of succinate, a gut bacterial metabolite regarded as undesirable when accumulated in the lumen, was significantly (p<0.05) higher in the female endurance runners. Faecalibacterium, that was significantly (p<0.05) abundant in female endurance runners, can produce succinate in certain environments and hence may contribute to succinate accumulation, at least partly. The present work suggested that the gut microbiotas of female endurance runners are seemingly dysbiotic when compared with those of control subjects. Further investigation of the mechanism by which intensive, prolonged exercise affects the gut microbiota is recommended.
Keywords: exercise-induced gastrointestinal disorders, gut microbiota, organic acids, putrefactive metabolites, female endurance runner
Introduction
Athletes are more predisposed to experience of gastrointestinal disorders after practices and competition, which is known as exercise-induced gastrointestinal syndrome.(1,2) Although frequency of occurrence of the gastrointestinal syndrome differs depending on various factors such as the type of sport, intensity/duration of the exercise and gender, typical symptoms often observed are diarrhea, loose stool, abdominal pain, and/or intestinal bleeding in the lower digestive tract.(2–4) However, the mechanism of this disorder are yet to be fully elucidated. According to Costa et al.,(2) two primary pathways are suggested as causatives of this disorder. One of such pathways is the circulatory-gastrointestinal pathway, in which prolonged high-intensity exercise causes gut ischemia-reperfusion that is the factor of site-specific oxidative stress and intestinal injury.(3,5) The other is the neuroendocrine-gastrointestinal pathway, in which both physical and psychological stresses alter gut motility and transit through enteric nervous activity, which consequently causes gut malabsorption of nutrients.(6,7) Indeed, previous work has showed that intense endurance running causes malabsorption of carbohydrates in triathletes.(8)
Changes in the gut microbiota caused by intensive exercise may also contribute to exercise-induced gastrointestinal disorders. For example, it has been reviewed elsewhere that gut dysbiosis, in other words, aberrant microbiota composition showing abnormal profiles of microbiota metabolites, can cause various gut disorders such as constipation, diarrhea and gut inflammation.(9,10)
In general, adequate exercise is regarded as beneficial for the gut.(11,12) For example, the abundances of butyrate-producing bacteria and concentrations of butyrate were observed to increase in humans after physical exercise.(13,14) However, this does not seem to be the case when the exercise is highly intensive and/or is physiologically stressful. Using a murine model, Allen et al.(15) reported that the effect on the gut microbiota is different when the exercise is voluntary or forced. Allen et al. found that voluntary exercise was beneficial for the gut health but forced exercise was not, because while the former attenuated gut inflammation, the latter further exacerbated it. Nonetheless, when compared with those of people living a regular lifestyle, the effect of highly-intensive exercise on the gut microbiotas of athletes remains unclear.(16,17)
In the present study, we aimed to obtain a basic understanding of the effect of highly-intensive endurance exercise on the gut microbiota for the possible development of future treatments of exercise-induced gastrointestinal disorders. We compared the gut environment, that is, the gut microbiota and its metabolites, between female elite endurance runners and non-athletic, but healthy women of similar age living within close geographic locations. In addition, fecal microbiota composition and three gut environmental parameters, namely the moisture content, the concentrations of organic acids and the concentrations of putrefactive metabolites were analyzed.
Materials and Methods
Ethics statements
The experiment was approved by the Ethical Committee of Kyoto Prefectural University and conducted as per their guidelines (approval number: 148). A written informed consent was obtained from all participants.
Research subjects
Freshly evacuated feces were collected from female Japanese elite endurance runners belonging to a company team [endurance runners (ER) group: n = 15, monthly running distance; 547 ± 111 km] and healthy female volunteers living in similar geographic location (control group: n = 14) (Table 1). All fecal specimens were collected from April 2018 to April 2019. There was no restriction on the timing and situation of fecal collection. The feces were collected using a fecal collection tube with a scoop (Sarstedt K.K., Tokyo, Japan) and immediately stored at –20°C or lower until further processing. It was not possible to analyze the fecal moisture content and the concentrations of putrefactive metabolites in one sample from the ER group because the fecal amount provided by the subject was too small. Therefore, for the analyses of fecal moisture content and putrefactive metabolites, the number of samples available from the ER group was 14.
Table 1.
Profiles of enrolled subjects
| Variable | Control | ER | p value |
|---|---|---|---|
| Total n (male/female) | 14 (0/14) | 15 (0/15) | |
| Age (years) | 20.9 ± 0.3 | 20.5 ± 1.2 | 0.80 |
| Self reported BMI | 21.3 ± 0.6 | 17.8 ± 0.2 | <0.01** |
**p<0.01.
Analysis of fecal microbiota by 16S rRNA metagenomics
Extraction of bacterial DNA from feces, library preparation and deep sequencing (MiSeq, Illumina K.K., Tokyo, Japan) were carried out exactly as described by Inoue et al.(18) Sequence data analysis was carried out as described by Hashimoto et al.(19) using QIIME 2.(20)
Measurement of the fecal moisture content
Fecal moisture contents were determined by lyophilization as described elsewhere.(21)
Analysis of the concentrations of fecal organic acids
The concentrations of fecal organic acids such as succinate, lactate, formate, acetate, propionate, iso-butyrate, n-butyrate, iso-valerate and n-valerate were measured by an ion-exclusion high-performance liquid chromatography as described elsewhere.(22)
Analysis of the concentrations of fecal putrefactive metabolites
Fecal putrefactive metabolites such as phenol, indole, skatole and para-cresol were measured by gas chromatography-mass spectrometry. Briefly, feces (100 mg) were thawed and transferred to 2 ml screw-cap tubes (Watson Co. Ltd., Tokyo, Japan) with a φ5.5 stainless steel ball (TOMY SEIKO Co. Ltd., Tokyo, Japan). Methanol (0.9 ml) and 10 µl of an internal standard methanol solution [1 mmol/L phenol-d5 (Sigma-Aldrich Japan, Tokyo, Japan)] were added and the feces were homogenized using Micro Smash MS-100 (3,000 r.p.m., 30 s). After centrifugation (15,000 × g; 5 min; 4°C), the supernatant was collected into a new 3.5 ml screw-capped cryotube (TPP Techno Plastic Products, Trasadingen, Switzerland). The residues were re-suspended in 1 ml of methanol, homogenized, centrifuged, and the supernatant was collected again as described above. This procedure was conducted two times. Total collected supernatants (approximately 2.9 ml) were mixed and used as samples for further analysis.
The method for the analysis of putrefactive metabolites was that described by GL Sciences Inc. (Tokyo, Japan; https://www.gls.co.jp/viewfile/?p=GA119). Briefly, the analysis was conducted using a gas chromatography mass spectrometry (GCMS-QP2010 Ultra; Shimadzu, Kyoto, Japan) equipped with an autosampler (AOC-5000; Shimadzu, Kyoto, Japan). Chromatographic separation was carried out with an InertCap5 column (30 m × 0.25 mm I.D. × 0.25 µm; GL Sciences Inc.). Helium was used as the carrier gas, and the linear velocity was set at 48.1 ml s. The stepwise thermal conditions of the column oven were as follows: the temperature was initially raised to and maintained at 40°C for 2 min; then it was raised to 320°C at a rate of 10°C/min and maintained for 10 min. The total run time of the analysis was 40 min. The mass spectrometer was set in selected ion monitoring mode at m/z 117.0 of indole, m/z 108.0 of para-cresol, m/z 94.0 of phenol, m/z 99.0 of phenol-d5, and m/z 131.0 of skatole. The injection was set in split mode (10.0) with an injection volume of 1 µl and an injector temperature of 250°C. The chemical ionization mode was set as follows: an ion source temperature of 200°C and an interface temperature of 250°C. The concentration of each parameter was calculated by the peak area of the parameter and the peak area of the internal standard (phenol-d5).
Statistical analysis
α-Diversity indices Chao1 (richness) and Shannon (evenness) were calculated with the R phyloseq package.(23) β-Diversity was estimated based on UniFrac distances by QIIME 1.9.1 and tested by a principal coordinate analysis (PCoA). The UniFrac distance between samples was statistically analyzed by a permutational multivariate analysis of variance (PERMANOVA) by QIIME 1.9.1. Age and BMI were statistically compared by the Welch’s t test. α-Diversity indices, the relative abundance (%) of bacterial phyla and genera, the fecal moisture content, and the concentrations of organic acids and putrefactive metabolites were statistically compared by the Wilcoxon’s rank sum test. Differences between the means were considered to be significant if p<0.05 and with a tendency to be significant if 0.05<p<0.1. Values are expressed as the means ± SE. All statistical analyses were calculated by R (ver. 3.6.0), unless otherwise specified.
Results
Diversity analysis of the gut microbiota
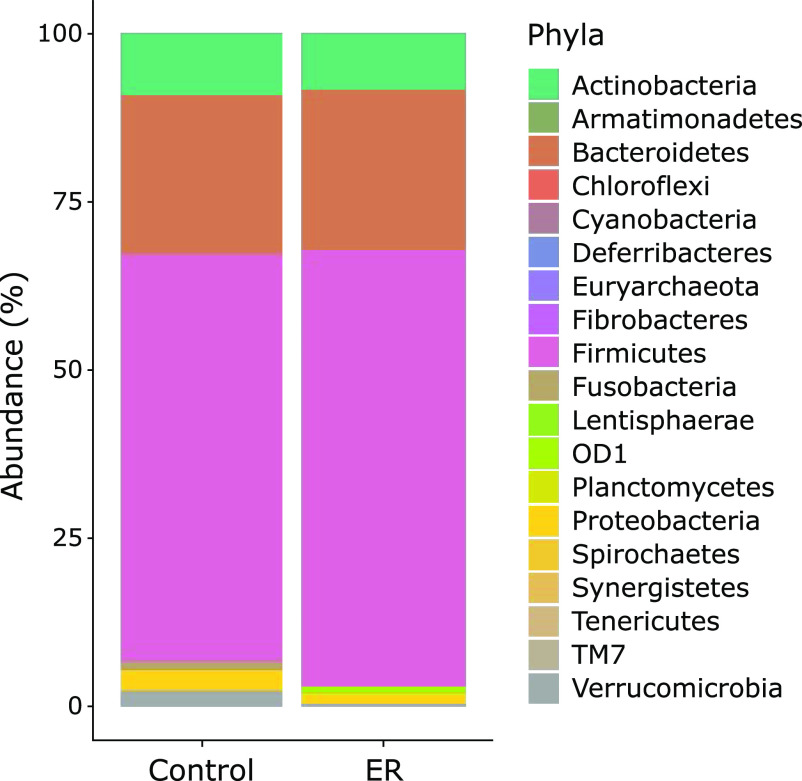
In the alpha-diversity analysis, the Chao1 index of the ER group showed a higher value (178 ± 13) than the control group (131 ± 35), although the difference was not significant (p = 0.09, Fig. 1). Similarly, no significant differences in the Shannon index were found between the two groups (p = 0.48). In contrast, the β-diversity between two groups was found to be significantly (p<0.05) different (Fig. 2). In the taxonomy analysis at the phylum level, the abundance of Deferribacteres was significantly higher in the ER group (0.006 ± 0.002%) than in the control group (0%). In addition, the abundance of OD-1 was also significantly higher in ER group (0.904 ± 0.167%) than in the control group (<0.001%) (Fig. 3). At the genus level, the relative abundances of 16 bacterial genera were significantly different between the two experimental groups (Table 2). For example, the abundance of 11 bacterial genera were significantly higher in the ER group than in the control group. In particular, the relative abundance of genera Faecalibacterium was more than 2-fold higher in the ER group (13.83 ± 1.55%) than in the control group (6.20 ± 1.14%).
Fig. 1.
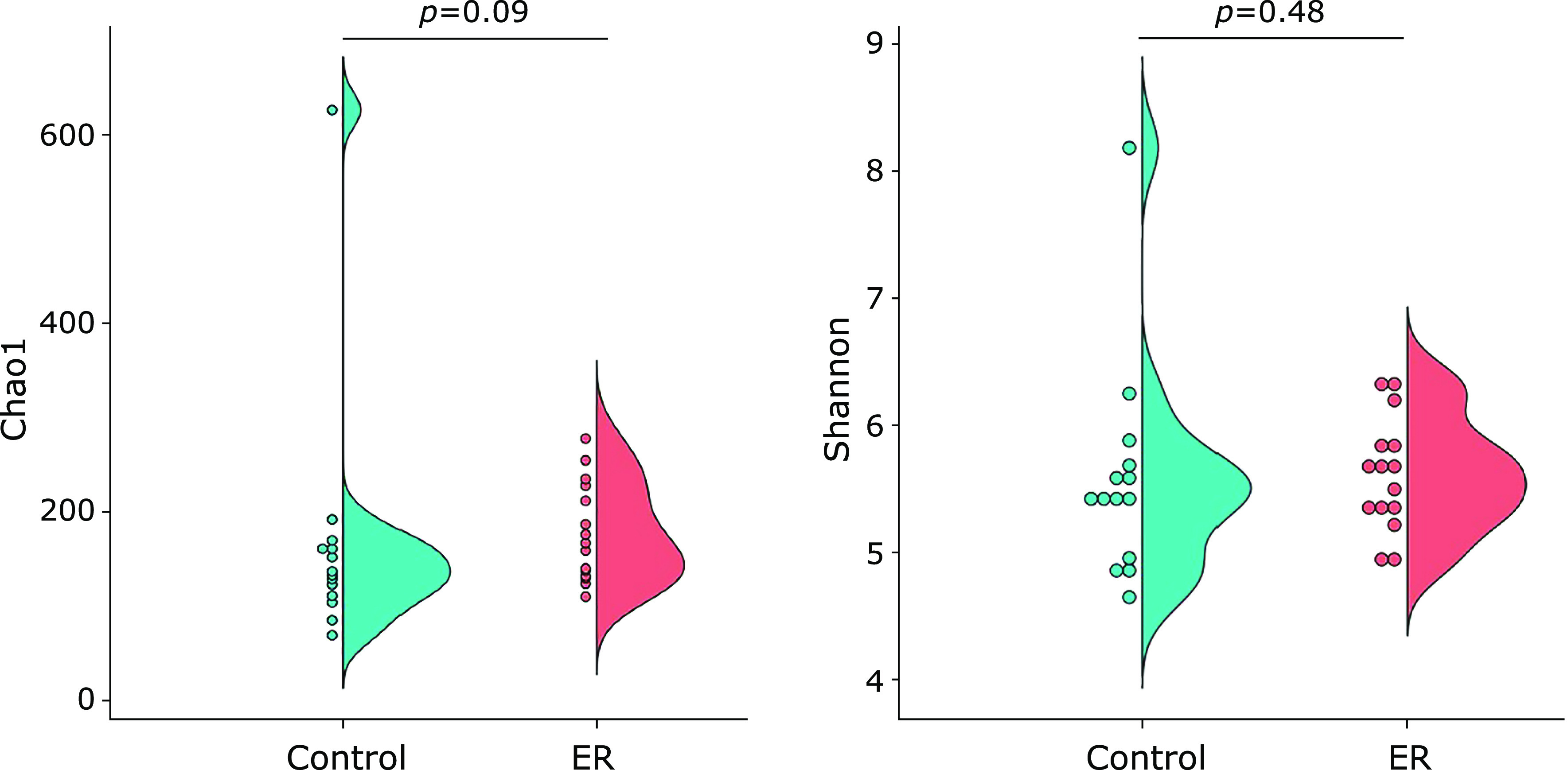
Chao1 and Shannon indices of the gut microbiota.
Fig. 2.
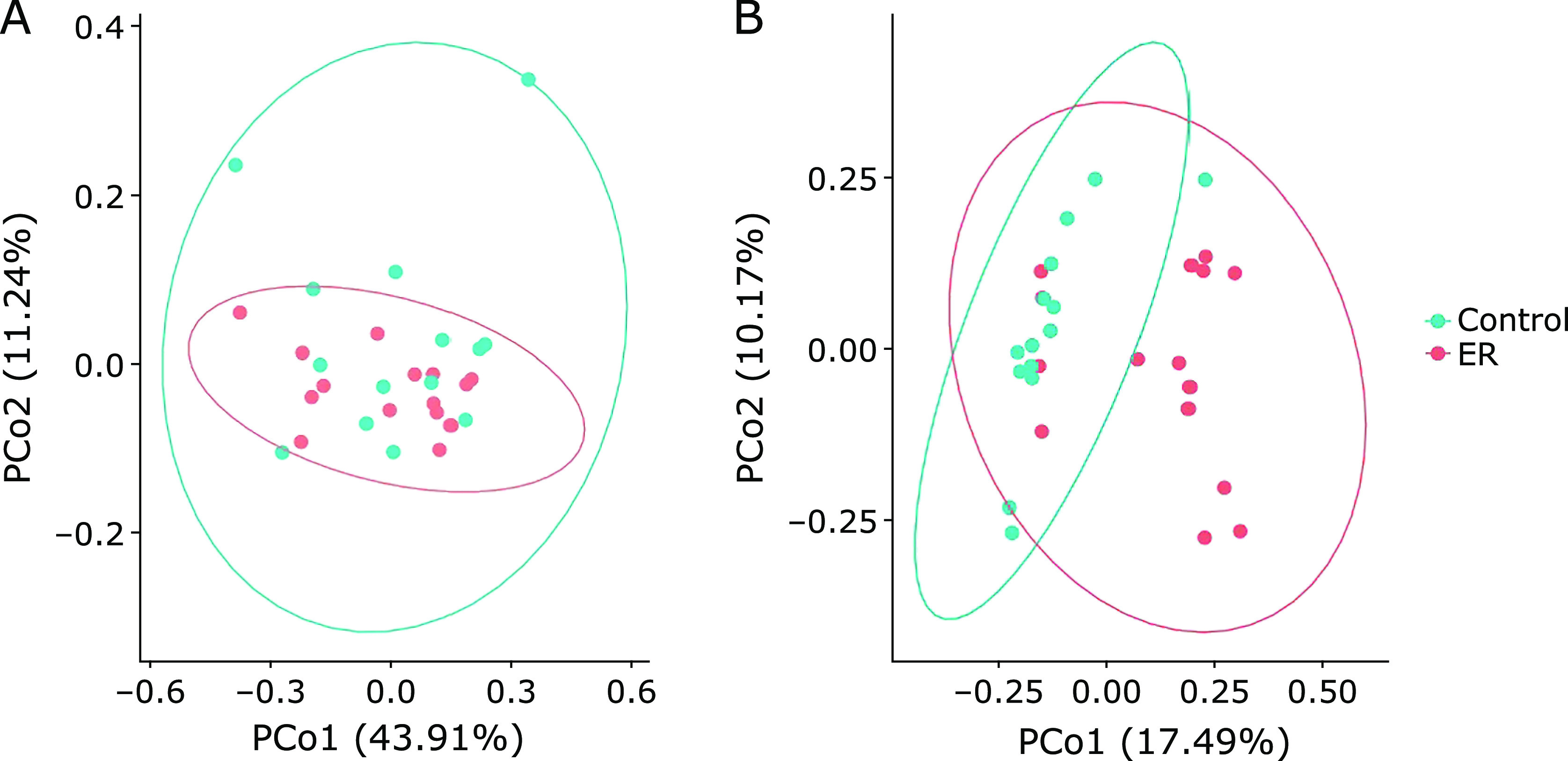
PCoA (principal coordinate analysis) based on weighted UniFrac (A) and unweighted UniFrac distances (B) generated from the data of the fecal microbiota. The ellipse enclosing each cluster indicate a 95% confidence interval. The PERMANOVA analysis indicated that the gut microbiota compositions of the ER and control groups were different, based on the unweighted UniFrac (p<0.05), but not on the weighted UniFrac distance.
Fig. 3.
The relative abundance of gut bacteria at the phylum level.
Table 2.
List of bacterial genus that significantly different abundance between control and ER group
| Nearest known bacterial genus | Abundance (%) |
||
|---|---|---|---|
| Control | ER | p value | |
| g__Acinetobacter | <0.01 | 0.01 ± 0.00 | 0.001** |
| g__Actinomyces | 0.16 ± 0.09 | 0.02 ± 0.01 | 0.004** |
| g__Bacillus | 0.01 ± 0.01 | 0.06 ± 0.02 | 0.037* |
| g__Bilophila | 0.18 ± 0.05 | 0.04 ± 0.03 | 0.033* |
| g__Citrobacter | 0.16 ± 0.14 | <0.01 | 0.032* |
| g__Clostridium | 0.32 ± 0.10 | 0.87 ± 0.17 | 0.017* |
| g__Faecalibacterium | 6.20 ± 1.14 | 13.83 ± 1.55 | 0.001** |
| g__Haemophilus | 0.02 ± 0.02 | 1.01 ± 0.40 | 0.008** |
| g__Lachnospira | 0.69 ± 0.36 | 1.94 ± 0.36 | 0.001** |
| g__Mucispirillum | <0.01 | 0.01 ± 0.00 | 0.005** |
| f__Paraprevotellaceae g__Prevotella | 0.13 ± 0.13 | 0.01 ± 0.00 | 0.004** |
| g__Rothia | <0.01 | 0.03 ± 0.01 | 0.001** |
| Unclassified p__OD1 | <0.01 | 0.90 ± 0.17 | <0.001** |
| Unclassified f__Clostridiaceae | <0.01 | 0.05 ± 0.04 | 0.022* |
| Unclassified f__Pasteurellaceae | <0.01 | 0.01 ± 0.01 | 0.045* |
| Unclassified f__S24-7 | 0.06 ± 0.06 | 0.02 ± 0.01 | 0.019* |
*p<0.05, **p<0.01.
The moisture content, and the concentrations of organic acids and putrefactive metabolites in feces (Table 3)
Table 3.
Moisture content, organic acid concentrations and putrefactive metabolites concentrations in the feces
| Measurements | Control | ER | p value |
|---|---|---|---|
| Moisture content (%) | 75.97 ± 2.32 | 76.62 ± 2.54 | 0.77 |
| Organic acids (mmol/kg of wet feces) | |||
| Succinate | 1.75 ± 1.08 | 3.56 ± 1.41 | 0.03* |
| Lactate | 0.06 ± 0.06 | 0.50 ± 0.32 | 0.29 |
| Formate | 0.05 ± 0.05 | 0.41 ± 0.19 | 0.15 |
| Acetate | 54.41 ± 4.06 | 64.89 ± 9.92 | 0.75 |
| Propionate | 19.61 ± 1.62 | 21.25 ± 3.43 | 0.59 |
| iso-Butyrate | 0.86 ± 0.37 | 2.60 ± 0.92 | 0.17 |
| n-Butyrate | 13.54 ± 1.46 | 15.34 ± 3.68 | 0.40 |
| iso-Valerate | 1.52 ± 0.39 | 2.71 ± 1.13 | 0.99 |
| n-Valerate | 0.84 ± 0.31 | 1.65 ± 0.68 | 0.73 |
| Total organic acid | 92.65 ± 6.46 | 115.63 ± 18.95 | 0.95 |
| Putrefactive metabolites (µg/g of wet feces) | |||
| Phenol | 4.65 ± 0.73 | 4.78 ± 0.54 | 0.56 |
| para-Cresol | 73.66 ± 15.63 | 85.57 ± 24.70 | 0.89 |
| Indole | 63.23 ± 13.58 | 38.95 ± 11.68 | 0.09 |
| Skatole | 6.43 ± 3.81 | 21.92 ± 14.67 | 0.87 |
*p<0.05.
There was no significant difference in the fecal moisture content between the two experimental groups. The concentration of fecal succinate was significantly higher in the ER group (3.56 ± 1.41 mmol/kg wet feces) than in the control group (1.75 ± 1.08 mmol/kg wet feces). The concentration of fecal indole tended to be lower (p = 0.09), in the ER group (38.95 ± 11.68 µg/g wet feces) than in the control group (63.23 ± 13.58 µg/g wet feces). As for the other organic acids and putrefactive metabolites, no significant differences in the concentrations were found between the two experimental groups.
Discussion
In the present work, we showed that gut microbiotas in the ER group, that is, the female elite endurance runners, were different from those in the control group. No significant differences were found in the Chao1 and Shannon indices between the two experimental groups (Fig. 1). However, one subject in the control group showed abnormally high values in both indices (Chao1: 627; Shannon: 8.18), which were regarded as outliers by the Smirnov-Grubbs test. When the values of this subject were excluded from the statistical comparison, the Chao1 index was significantly (p = 0.03) higher in the ER group (178 ± 13) than in the control group (133 ± 4). Therefore, we believed that the ER group may have had a higher richness in their gut microbiotas than did the control group. Nonetheless, this significance was not observed in the Shannon index even after excluding the outlying value (p = 0.27). Separately, the PCoA based on the unweighted UniFrac distance, also showed that the composition of the gut microbiotas in the ER group were different than those in control group (Fig. 2; PERMANOVA; p<0.05).
The taxonomy analysis detected that some bacteria, which have been suggested as being associated with gut inflammation, were more abundant in the ER group than in the control group. For instance, genus Haemophilus and Rothia were reported to be abundant in Crohn’s disease patients.(24) In addition, Mucispirillum, a mucus degradation bacterium, was found to be abundant in a murine inflammation model.(25,26) Similarly, Ruminococcus gnavus,(27) another mucus degradation bacterium, is reported to increase in the gut of mice when exercise was forcibly imposed, but not when it was voluntarily done by the animals.(15) Therefore, intensive exercise likely affects the production and/or sugar-chain composition of mucus in the gut.
By contrast, Faecalibacterium, which was also significantly (p<0.05) abundant in the ER group, is a butyrate-producing bacterium that is generally known to be beneficial for the host’s health,(28,29) especially for its anti-inflammatory effect on the gut.(30) Therefore, in the present work, a higher abundance of Faecalibacterium was seemingly unlikely when possible proinflammatory bacteria such as Mucispirillum and family Pasteurellaceae were also present in high abundance, if a gut inflammation status is considered. However, Hansen et al.(31) pointed out that Faecalibacterium may have a more complex role in IBD (inflammatory bowel disease) that goes beyond our current understanding, because they found a high abundance of Faecalibacterium in biopsy samples of inflamed tissues from IBD children. Moreover, in the present study, a high abundance of Faecalibacterium may have not necessarily been acting as a health-promoting bacterium, but instead, as an opportunistic microorganism, due to the altered gut environment of the ER group.
It is worth noting that Faecalibacterium is known for its succinate production. Indeed, Faecalibacterium produces succinate in gut environments rich in fumarate, for instance.(32) Under normal conditions, the succinate produced by gut bacteria is quickly metabolized to propionate, but it can accumulate under abnormal conditions.(33) For example, some Bacteroides species such as B. fragilis produce propionate via succinate, but this pathway does not work well in an environment with excessive carbohydrates or lack of vitamin B12. As a result, succinate is not metabolized and accumulated.(34,35) In the present study, a plausive explanation for the significantly higher concentration of succinate may be due to the abnormal metabolic pathway of Faecalibacterium in the microbiota of the ER group, probably affected by the intensive, prolonged exercise. It has been suggested that exercise increases the abundances of butyrate-producing bacteria such as Faecalibacterium and Rosebuira in human subjects.(13,14) Although, our results showed a high abundance of Faecalibacterium, which is in agreement with previous work, no increase in butyrate was observed. This apparent discrepancy may be explained by the fact that the abnormal gut environment of the ER group led Faecalibacterium to produce succinate, but not butyrate.
The accumulation of succinate seems to increase the osmotic pressure in the lumen and to reduce the water absorption rate of the intestine.(36) Therefore, succinate accumulation may be a risk factor for diarrhea or loose stool. In addition, the association of succinate accumulation with gut inflammation have been previously reported.(37) Therefore, at least partly, succinate accumulation is likely a causative factor of exercise-induced gastrointestinal disorders.
The analysis of fecal putrefactive metabolites showed that the concentration of indole tended to be lower in the ER group (Table 3). The concentration of the putrefactive metabolites in the feces generally counteract the moisture content and the concentrations of organic acids.(38) Thus, we theorize that, unlike that of the control group, the gut environment of the ER group was prone to produce more organic acids than putrefactive metabolites.
The prolonged, high-intensity exercise that endurance runners routinely carry out may have been resulted in a form of dysbiosis. For example, the production of reactive oxygen species and inflammatory cytokines associated with gut ischemia-reperfusion may have been caused by this kind of exercise, which may have negatively affected the gut microbiota due to their bactericidal properties.(1,5,39) The exercise also likely caused gut injury and malabsorption of nutrients. Nonetheless, the underlying mechanism by which intensive, prolonged exercise affects the gut microbiota modification still remains unclear.
To conclude, in the present study, we showed that female elite endurance runners had gut microbiotas and metabolites profiles that differed from those observed in age-matching female controls. In addition, a higher abundance of Faecalibacterium in the gut microbiota of the female elite endurance runners may be involved in the accumulation of succinate. We theorize that a higher abundance of inflammation-related bacteria and a higher concentration of succinate may have resulted in a form of dysbiosis in the athletes. We suggest that the relationship between the microbiota and the production of bacterial metabolites in the gut of athletes be further evaluated in future studies.
Author Contributions
WA, TTakagi, YN, and RI conceived this experiment; WA and AK enrolled the participating subjects and collected fecal samples; SM, TK, and RI conducted the experiments; SM, TTsukahara, and RI analyzed data; SM, TTsukahara, and RI wrote the manuscript. All authors have read and approved the final manuscript.
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Clark A, Mach N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J Int Soc Sports Nutr 2016; 13: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa RJS, Snipe RMJ, Kitic CM, Gibson PR. Systematic review: exercise-induced gastrointestinal syndrome-implications for health and intestinal disease. Aliment Pharmacol Ther 2017; 46: 246–265. [DOI] [PubMed] [Google Scholar]
- 3.ter Steege RWF, Kolkman JJ. Review article: the pathophysiology and management of gastrointestinal symptoms during physical exercise, and the role of splanchnic blood flow. Aliment Pharmacol Ther 2012; 35: 516–528. [DOI] [PubMed] [Google Scholar]
- 4.Halvorsen FA, Lyng J, Glomsaker T, Ritland S. Gastrointestinal disturbances in marathon runners. Br J Sports Med 1990; 24: 266–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadatani Y, Watanabe T, Shimada S, Otani K, Tanigawa T, Fujiwara Y. Microbiome and intestinal ischemia/reperfusion injury. J Clin Biochem Nutr 2018; 63: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leiper JB, Nicholas CW, Ali A, Williams C, Maughan RJ. The effect of intermittent high-intensity running on gastric emptying of fluids in man. Med Sci Sports Exerc 2005; 37: 240–247. [DOI] [PubMed] [Google Scholar]
- 7.van Wijck K, Pennings B, van Bijnen AA, et al. Dietary protein digestion and absorption are impaired during acute postexercise recovery in young men. Am J Physiol Regul Integr Comp Physiol 2013; 304: R356–R361. [DOI] [PubMed] [Google Scholar]
- 8.Peters HPF, Schep G, Koster DJ, Douwes AC, de Vries WR. Hydrogen breath test as a simple noninvasive method for evaluation of carbohydrate malabsorption during exercise. Eur J Appl Physiol Occup Physiol 1994; 68: 435–440. [DOI] [PubMed] [Google Scholar]
- 9.Ohkusa T, Koido S, Nishikawa Y, Sato N. Gut microbiota and chronic constipation: a review and update. Front Med (Lausanne) 2019; 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barko PC, McMichael MA, Swanson KS, Williams DA. The gastrointestinal microbiome: a review. J Vet Intern Med 2018; 32: 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gubert C, Kong G, Renoir T, Hannan AJ. Exercise, diet and stress as modulators of gut microbiota: implications for neurodegenerative diseases. Neurobiol Dis 2020; 134: 104621. [DOI] [PubMed] [Google Scholar]
- 12.Codella R, Luzi L, Terruzzi I. Exercise has the guts: how physical activity may positively modulate gut microbiota in chronic and immune-based diseases. Dig Liver Dis 2018; 50: 331–341. [DOI] [PubMed] [Google Scholar]
- 13.Bressa C, Bailén-Andrino M, Pérez-Santiago J, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One 2017; 12: e0171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen JM, Mailing LJ, Niemiro GM, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc 2018; 50: 747–757. [DOI] [PubMed] [Google Scholar]
- 15.Allen JM, Berg Miller ME, Pence BD, et al. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol (1985) 2015; 118: 1059–1066. [DOI] [PubMed] [Google Scholar]
- 16.O’Sullivan O, Cronin O, Clarke SF, et al. Exercise and the microbiota. Gut Microbes 2015; 6: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mach N, Fuster-Botella D. Endurance exercise and gut microbiota: a review. J Sport Health Sci 2017; 6: 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue R, Ohue-Kitano R, Tsukahara T, et al. Prediction of functional profiles of gut microbiota from 16S rRNA metagenomic data provides a more robust evaluation of gut dysbiosis occurring in Japanese type 2 diabetic patients. J Clin Biochem Nutr 2017; 61: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto Y, Hamaguchi M, Kaji A, et al. Intake of sucrose affects gut dysbiosis in patients with type 2 diabetes. J Diabetes Investig 2020. DOI: 10.1111/jdi.13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019; 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshi S, Sakata T, Mikuni K, Hashimoto H, Kimura S. Galactosylsucrose and xylosylfructoside alter digestive tract size and concentrations of cecal organic acids in rats fed diets containing cholesterol and cholic acid. J Nutr 1994; 124: 52–60. [DOI] [PubMed] [Google Scholar]
- 22.Tsukahara T, Matsukawa N, Tomonaga S, Inoue R, Ushida K, Ochiai K. High-sensitivity detection of short-chain fatty acids in porcine ileal, cecal, portal and abdominal blood by gas chromatography-mass spectrometry. Anim Sci J 2014; 85: 494–498. [DOI] [PubMed] [Google Scholar]
- 23.McMurdie PJ, Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac Symp Biocomput 2012; 235–246. [PMC free article] [PubMed] [Google Scholar]
- 24.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014; 15: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan I, Ullah N, Zha L, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019; 8: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loy A, Pfann C, Steinberger M, et al. Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2017; 2: e00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Getachew B, Aubee JI, Schottenfeld RS, Csoka AB, Thompson KM, Tizabi Y. Ketamine interactions with gut-microbiota in rats: relevance to its antidepressant and anti-inflammatory properties. BMC Microbiol 2018; 18: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miquel S, Martín R, Bridonneau C, et al. Ecology and metabolism of the beneficial intestinal commensal bacterium Faecalibacterium prausnitzii. Gut Microbes 2014; 5: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol 2014; 6: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Säemann MD, Böhmig GA, Österreicher CH, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J 2000; 14: 2380–2382. [DOI] [PubMed] [Google Scholar]
- 31.Hansen R, Russell RK, Reiff C, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohnʼs but not in ulcerative colitis. Am J Gastroenterol 2012; 107: 1913–1922. [DOI] [PubMed] [Google Scholar]
- 32.Khan MT, Duncan SH, Stams AJ, van Dijl JM, Flint HJ, Harmsen HJ. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J 2012; 6: 1578–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichardt N, Duncan SH, Young P, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 2014; 8: 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc 2003; 62: 67–72. [DOI] [PubMed] [Google Scholar]
- 35.Chen M, Wolin MJ. Influence of heme and vitamin B12 on growth and fermentations of Bacteroides species. J Bacteriol 1981; 145: 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukahara T, Ushida K. Succinate accumulation in pig large intestine during antibiotic-associated diarrhea and the constitution of succinate-producing flora. J Gen Appl Microbiol 2002; 48: 143–154. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Veledo S, Vendrell J. Gut microbiota-derived succinate: friend or foe in human metabolic diseases? Rev Endocr Metab Disord 2019; 20: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda N, Saito Y, Shimizu J, Ochi A, Mizutani J, Watabe J. Variations in concentrations of bacterial metabolites, enzyme activities, moisture, pH and bacterial composition between and within individuals in faeces of seven healthy adults. J Appl Bacteriol 1994; 77: 185–194. [DOI] [PubMed] [Google Scholar]
- 39.Vatansever F, de Melo WC, Avci P, et al. Antimicrobial strategies centered around reactive oxygen species—bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol Rev 2013; 37: 955–989. [DOI] [PMC free article] [PubMed] [Google Scholar]