Fig. 2.
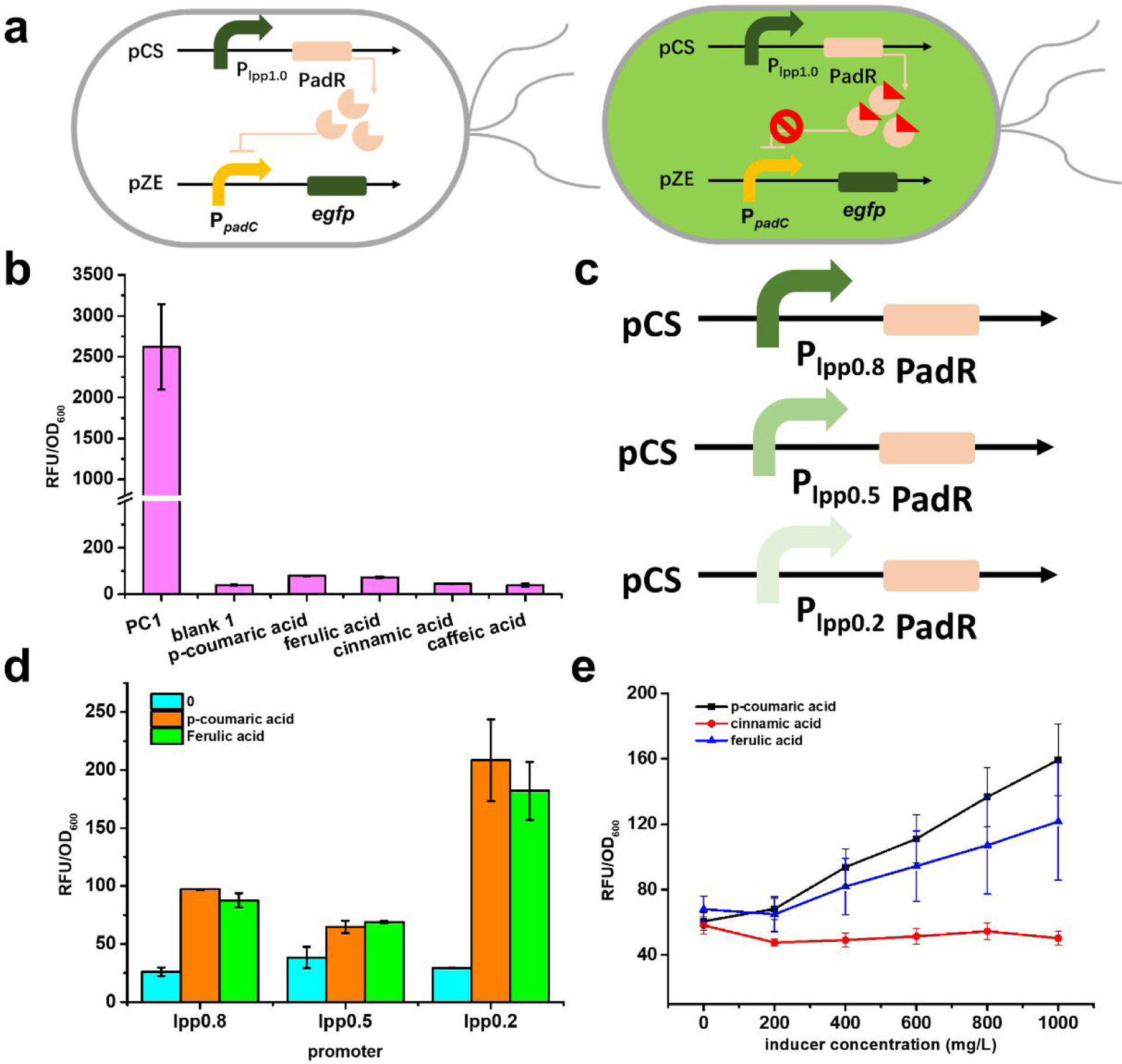
The design of PadR-PpadC biosensor system and its optimization. (a) The genetic circuit of PadR-PpadC biosensor system. There are two modules in this system, reporter module and regulator module. In the absence of p-coumaric acid, PadR can repress the PpadC and egfp cannot express successfully. When p-coumaric acid was added, it can bind to PadR, which released the inhibition of PpadC; the green fluorescence can be detected. (b) The promoter strength with or without PadR inhibition. PC1, without regulator module, was used to test the strength of PpadC; blank, with reporter module and regulator module, which was used to characterize the inhibition efficiency; p-coumaric acid, ferulic acid, and cinnamic acid represent that different inducers were added to release the PadR inhibition. (c) Different inducers in different concentration. P-coumaric acid, ferulic acid, and cinnamic acid were used as inducers to release the inhibition by pCS-lpp1.0-PadR. (d) Differe nt regulator modules with various promoter. (e) The effects of different expression level of PadR in decreasing its inhibition activity. All data are reported as mean±s.d. from three independent experiments (n=3). Error bars are defined as s.d.
