Extended Data Fig. 4 |. Sequence and structure requirements for deposition of ac4C.
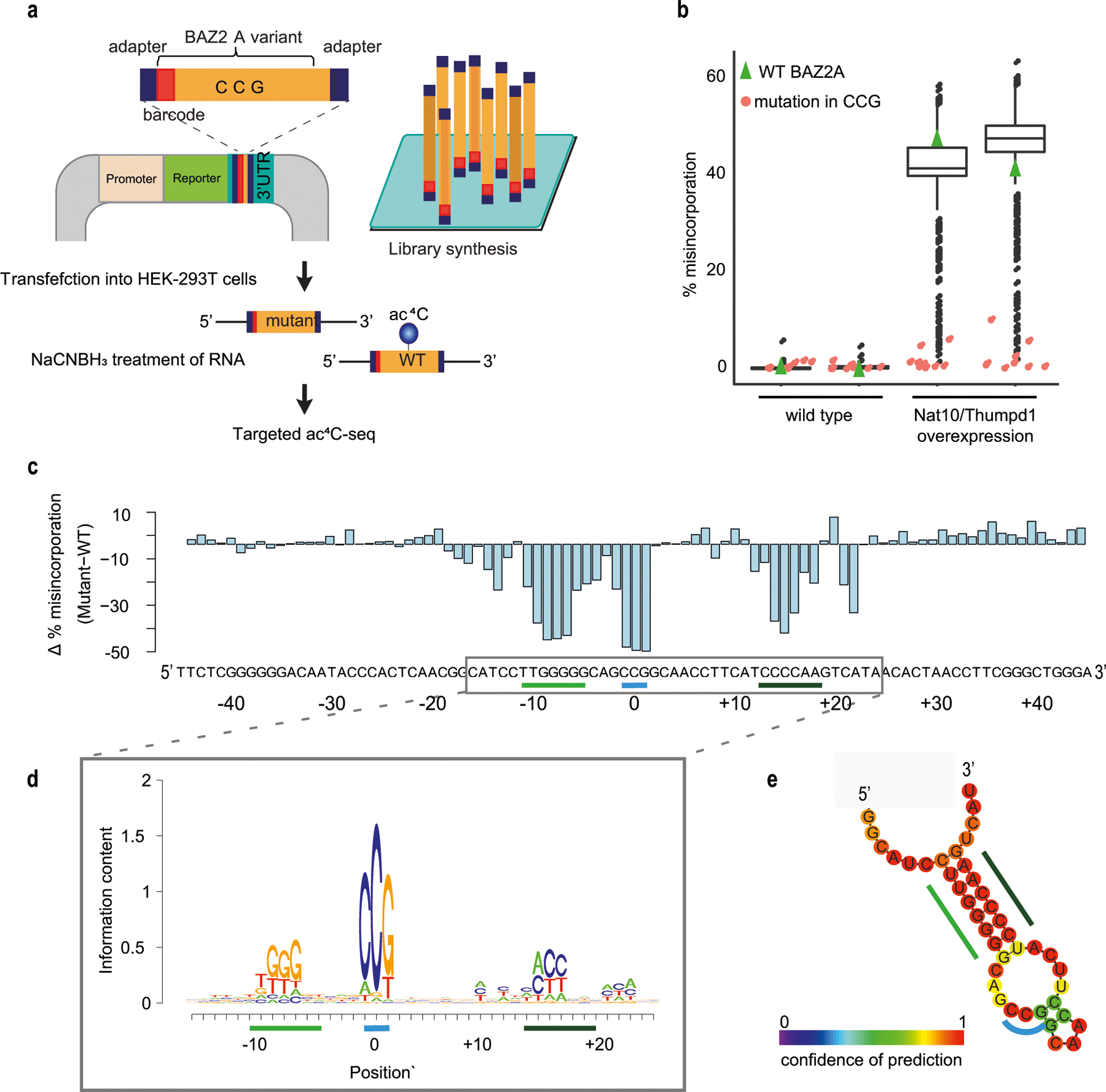
a, Oligonucleotides representing the wild-type sequence surrounding the acetylated site in BAZ2A mRNA, or variants with single mutations across the wild-type sequence, were synthesized as a pool and cloned into the 3′UTR of a reporter gene. The pool of plasmids was transfected into wild-type HEK-293T cells or cells transiently overexpressing NAT10 and THUMPD1. RNA extracted from cells was subjected to targeted ac4C-seq and ac4C levels were estimated on the basis of misincorporation rates. b, Misincorporation rate of oligonucleotides described in a, harbouring the wild-type sequence of BAZ2A (green triangles) or a sequence mutated at the CCG motif and at its surrounding bases (red and black, respectively). Box plot visualization parameters are as in Fig. 1h. n = 2 biologically independent samples. c, The difference in misincorporation rate of oligonucleotides with a single base mutation compared with the wild-type oligonucleotide is shown across all positions of the construct. d, De novo construction of the motifs surrounding the modified cytidine were built on the basis of the contribution of single-base mutations in the BAZ2A sequence to the reduction in misincorporation rate compared to wild-type BAZ2A sequence. e, Secondary structure of the BAZ2A mRNA fragment as predicted by RNAfold. Bases are colour-coded according to confidence level of the prediction. Regions highlighted by a blue and green line in c–e represent the CCG motif and a stem structure surrounding the modified cytidine, respectively.
