Abstract
Background
Antimonial drugs have long been the mainstay to treat visceral leishmaniasis. Their use has been discontinued in the Indian subcontinent because of drug resistance, but they are still clinically useful elsewhere. The goal of this study was to find markers of antimony resistance in Leishmania donovani clinical isolates and validate experimentally their role in resistance.
Methods
The genomes of sensitive and antimony-resistant clinical isolates were sequenced. The role of a specific gene in contributing to resistance was studied by CRISPR-Cas9–mediated gene editing and intracellular drug sensitivity assays.
Results
Both gene copy number variations and single nucleotide variants were associated with antimony resistance. A homozygous insertion of 2 nucleotides was found in the gene coding for the aquaglyceroporin AQP1 in both resistant isolates. Restoring the wild-type AQP1 open reading frame re-sensitized the 2 independent resistant isolates to antimonials. Alternatively, editing the genome of a sensitive isolate by incorporating the 2-nucleotide insertion in its AQP1 gene led to antimony-resistant parasites.
Conclusions
Through genomic analysis and CRISPR-Cas9–mediated genome editing we have proven the role of the AQP1 mutations in antimony clinical resistance in L. donovani.
Keywords: Leishmania, drug resistance, single nucleotide variants, copy number variation
This study reports on the genomics characterization of antimony-resistant Leishmania donovani clinical isolates and on the validation of the aquaglyceroporin AQP1 as a proven molecular marker for antimony resistance through CRISPR Cas9–mediated genome editing.
Visceral leishmaniasis (VL) is caused by Leishmania donovani, with 0.2–0.4 million new cases per year [1]. There are no human vaccines and chemotherapy is often the only option against VL. There are few licensed drugs and the mainstay has long relied on the antiquated pentavalent antimonials (SbV). Antimonials are still useful in parts of the world but not on the Indian subcontinent, the region with the highest rate of VL. Indeed, resistance to SbV in L. donovani is now widespread in India [2, 3]. Many factors were associated with resistance: exclusive anthroponotic cycle, suboptimal use or quality of drugs, or the contamination of drinking water by arsenic [4]. Indeed, Leishmania cells selected for resistance in vitro to either antimonite (SbIII) or arsenite (AsIII) were cross-resistant to one another [5]. Both SbIII and AsIII share the same transport system in Leishmania [6, 7]. The SbV drug is thought to be a prodrug that is reduced in the host macrophage or the parasite or both [3] to SbIII, the active form of the drug.
The search for molecular markers of resistance to antimonials has been exhaustive, with more than 20 genes that have been associated with antimonial resistance in vitro [3]. Three of those markers appear to be frequently associated with clinical resistance. One is the ABC transporter MRPA that produces resistance by sequestering the metal as a thiol conjugate into a vacuole [8]. The MRPA gene was found amplified [9, 10] or overexpressed [11, 12] in L. donovani clinical isolates resistant to SbV. Consistent with MRPA being an antimony-thiol ABC transporter, increased levels of thiols were observed in Leishmania selected in vitro for metal resistance [13, 14]. These thiols were increased by amplification and/or overexpression of genes involved in the synthesis of the building blocks of the glutathione-spermidine conjugate trypanothione [15]. Overexpression of genes involved in trypanothione biosynthesis was observed in antimony-resistant L. donovani clinical isolates [9, 11, 12, 16, 17]. The third marker found in clinical isolates is the aquaglyceroporin AQP1, a channel involved in the transport of SbIII and AsIII in Leishmania [7, 18]. Downregulation of AQP1 was found to correlate with antimony resistance in L. donovani [19, 20], although with some exceptions [12, 21].
Genome sequencing is now used routinely for studying resistance in Leishmania [22]. Two large sequencing studies were conducted with L. donovani–resistant isolates. One was helpful in revealing genomic structure and copy number variation (CNV) but less informative regarding markers of resistance [23]. The other reported a small insertion deletion (indel) in AQP1 that was found in cells resistant to antimonials [24]. Its role in resistance remains to be established, however. The same study [24] also reported an increased copy number of MRPA. In the present study we sequenced L. donovani–resistant clinical isolates, found similar indels in AQP1 along with known and new CNVs, and have validated by CRISPR-Cas9–mediated genome editing that the indel in AQP1 is indeed responsible for SbV resistance in L. donovani clinical isolates.
METHODS
Leishmania Parasites
Leishmania donovani FDA 9551 (9551), FDA 9515 (9515), and FDA 9518 (9518) isolates were isolated between 1995 and 1998 [2] and were grown in SDM-79 medium at 25°C supplemented with 10% fetal bovine serum and 5 μg/mL of hemin at pH 7. Strain 9551 was sensitive to antimony, whereas strains 9515 and 9518 were resistant [2].
Susceptibility Testing in Intracellular Macrophages
THP-1 cells (American Type Culture Collection T1B-202) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 IU penicillin/mL, and 100 μg streptomycin/mL at 37°C in a 5% CO2 atmosphere. THP-1 cells were differentiated for 2 days in RPMI 1640 medium containing 20 ng/mL of phorbol myristate acetate (PMA), as described [25]. Stationary-phase parasites, transfected with a firefly luciferase-containing vector [25], were used to infect PMA-differentiated THP-1 at a ratio of 25:1 (2 × 105 THP-1 cells/well), for 2 to 3 hours at 37°C. Extracellular parasites were washed and infected cells were maintained in the absence of drug for 48 hours, after which they were treated with SbV (sodium stibogluconate; Calbiochem) for 96 hours. Luciferase activity was measured [25] and half maximal effective concentration (EC50) values were calculated using GraphPad Prism 5.01 software. An average of at least 2 biological replicates each containing 4 technical replicates were performed.
Whole-genome Sequencing
Paired-ends sequencing libraries were prepared with the Nextera DNA sample prep kit and sequenced on an Illumina HiSeq platform. Sequence reads were aligned to the L. donovani BPK282A1 genome [23] using the software bwa-mem [26]. The maximum number of mismatches was 4, the seed length was 32, and 2 mismatches were allowed within the seed. Read duplicates were marked using Picard (http://broadinstitute.github.io/picard), and we applied the Genome Analysis Toolkit (GATK) for single nucleotides variants (SNVs) and indel discovery [27]. Copy numbers variations were derived from read depth coverage along small nonoverlapping genomic windows (5 kb) for the 36 chromosomes, as described [28]. Several python and bash scripts were created to further analyze the data.
Phylogenetics Analysis
Sequencing reads from a set of 52 samples (listed in Supplementary Table 1) [24] were downloaded from the Sequence Reads Archive (SRA) database. Sequence reads were aligned to the L. donovani BPK282A1 genome using bwa-mem [26]. Read duplicates were marked using Picard. Genome Variant Call Format (GVCF) files were generated for each sample using the HaplotypeCaller from GATK [27] version 4.1.2.0. GVCF files were merged then genotyped using the CombineGVCFs and GenotypeGVCFs tools from GATK version 4.1.2.0, respectively. The SNVs from the resulting Variant Call Format (VCF) file were filtered and converted into the phylip format using vcf2phylip version 2.0. Genome-wide phylogeny was computed with RAxML-NG [29] using the GTR+G substitution model and 500 bootstrap replicates. The phylogenetic network was visualized using SplitsTree5 [30].
DNA Editing
We used CRISPR-Cas9–mediated gene editing to remove the dinucleotide “TC” insertion in AQP1 in resistant isolates or to introduce it in the AQP1 gene of a sensitive isolate. This editing was selected by co-transfection of a gene cassette encoding a puromycin acetyltransferase (PURO) targeting the pteridine reductase 1 gene PTR1. This co-transfection strategy was used successfully for editing several genes involved in drug resistance in Leishmania [31]. Cells growing in the presence of puromycin were reputed to have their AQP1 gene edited, and this was confirmed by sequencing.
The targeting CRISPR RNAs (crRNAs) were designed using the Eukaryotic Pathogen CRISPR guide RNA (gRNA) Design Tool (http://grna.ctegd.uga.edu) [32] and synthetized by the Alt-R CRISPR service (Integrated DNA Technologies, USA), along with the universal trans-activating CRISPR RNA (tracrRNA). All primers sequences and gRNAs used in this study are listed in Supplementary Table 2. The TC insertion allowed the gRNA to target the mutated but not the wild-type AQP1 allele. This was exploited for the removal of the TC insertion from the resistant parasites, by transfecting these with the gRNA and a 600-bp repair cassette corresponding to the wild-type AQP1 sequence. To edit antimony-sensitive parasites, we relied on a gRNA directing Cas9 to cut in the vicinity of the TC insertion site that was co-transfected with a 120-bp repair cassette harboring the TC insertion along with a GCC to GCG synonymous mutation downstream to the Cas9 cleavage site for preventing Cas9 to cleave the edited sequence.
The crRNA:tracrRNA complex was formed according to the manufacturer’s instructions and co-transfected along with the AQP1 polymerase chain reaction (PCR) fragments and the PURO cassette targeting PTR1 in L. donovani strains expressing Cas9 [31]. Electroporated cells were incubated at 25°C for 24 hours, after which transfectants were selected with puromycin at a final concentration of 50–100 μg/mL. Clones were isolated on SDM-79 plates, resuspended in liquid SDM-79, and incubated at 25°C. Genomic DNA was purified and submitted to Sanger sequencing to verify the editing of the AQP1 locus.
Quantitative Real-time Polymerase Chain Reaction
The synthesis of the cDNA and real-time quantitative PCR were performed in triplicate, as previously described [33] using SYBR Green Supermix (BioRad) for detection in a Rotor Gene-3000 (Corbett Research). With an initial denaturation at 95°C for 4 minutes followed by 40 cycles with denaturation at 95°C for 20 seconds, annealing at 57° for 20 seconds, and extension at 72°C for 20 s. The amount of PCR products was determined using the relative standard curve method. The standard growth curves parameters were R2 ≥ 0.98; slope (M) at −3.3, and efficiency between 90% and 110%. They were normalized to the amount of the endogenous control gene, GAPDH.
RESULTS
Characterization of Leishmania donovani Clinical Isolates
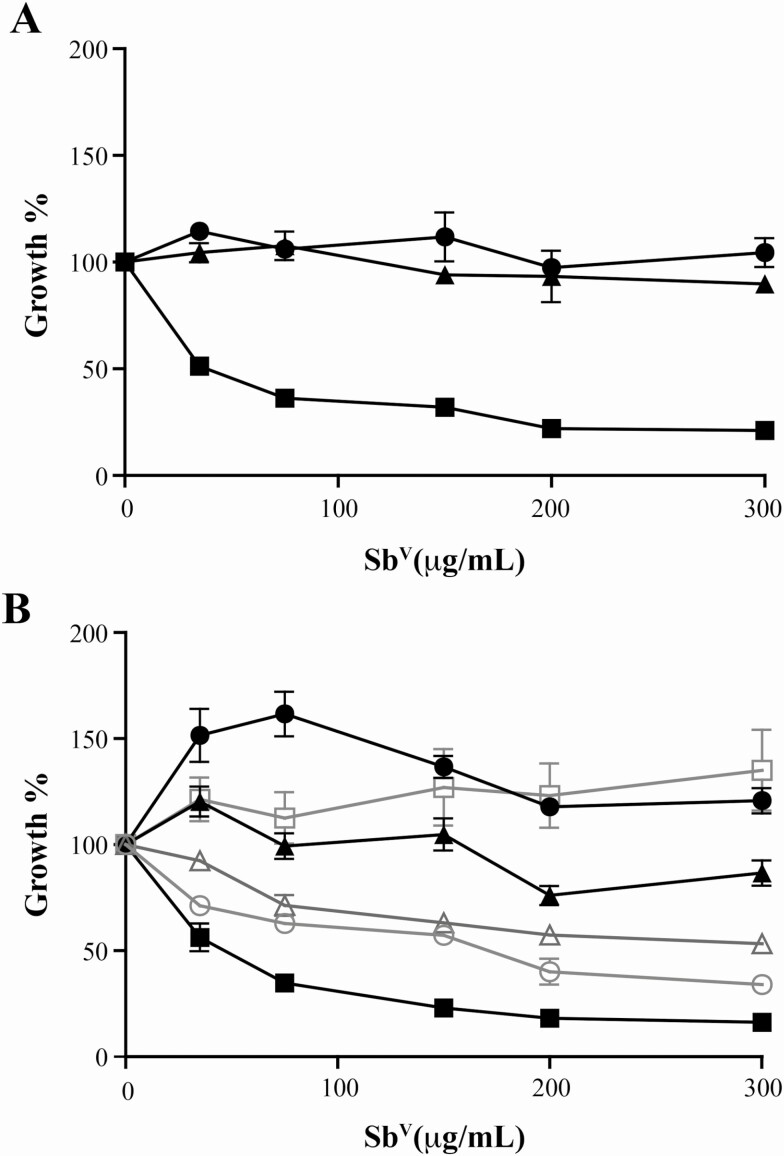
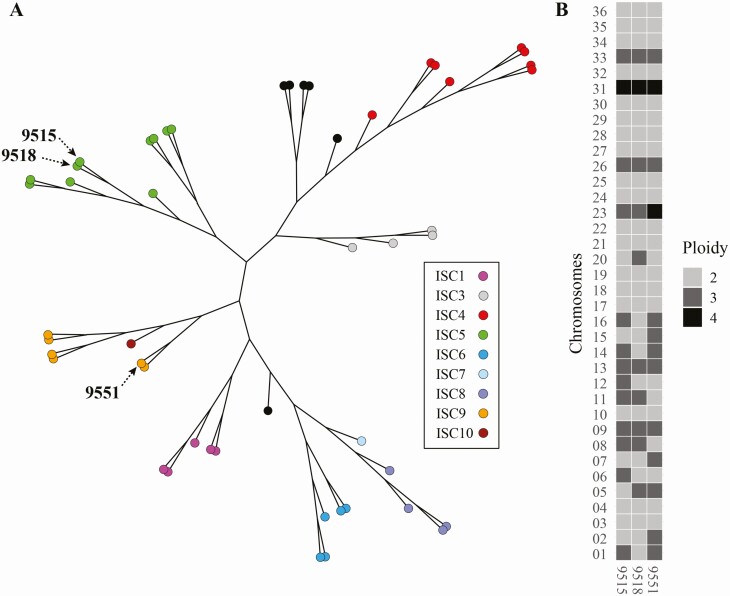
We confirmed using our intracellular macrophage assay that 9551 is sensitive to SbV, while 9515 and 9518, derived from patients who did not respond to chemotherapy, are resistant (Figure 1A). The genomic DNAs of the 3 clinical isolates were sequenced. Genome coverage for the 3 strains varied from 62- to 73-fold. These strains were isolated in the 1990s, a decade earlier than the strains sequenced under a large sequencing campaign [24]. A phylogenetic analysis of the 3 isolates with a subset of published L. donovani strains [24] indicated that they were clustering with the main core group of Indian L. donovani isolates (Figure 2A). The 2 resistant strains clustered with the core group ISC5 using the nomenclature of Imamura et al [24], while the sensitive strain 9551 clustered with the ISC9 group (Figure 2A). Read depth coverage over the 36 chromosomes of L. donovani was studied to predict CNVs. Half the chromosomes were diploid in the 3 isolates, 6 chromosomes were aneuploid in the 3 strains, and strain-specific variation in ploidy was observed for the 12 remaining chromosomes (Figure 2B). Polyploidy for chromosome 8 and 11 was specific to both resistant isolates while polyploidy for chromosome 2 and 7 was specific to the sensitive isolate.
Figure 1.
Susceptibility testing of pentavalent antimony in Leishmania donovani clinical isolates and their recombinants. A, Clinical isolates L. donovani 9515 (▲), 9518 (●), and 9551 (■) were transfected with the firefly luciferase [25] and used to infect THP-1 cells and treated with increasing concentrations of SbV. The average of 4 replicates is shown. All concentration points were statistically significant according to an unpaired t test (2-tailed) at P ≤ .001. B, The luciferase-expressing strains were transfected with a PURO PTR1 inactivation cassette and these parasites were used as controls (black lines for L. donovani 9515 [▲], 9518 [●], and 9551 [■]). The dinucleotide TC indels in AQP1 of 9515 (∆) and 9518 (○) were removed or inserted in 9551 (□) by genome editing, and their susceptibility to SbV is shown with gray lines. All concentration points were statistically significant using an unpaired t test (2-tailed) at P ≤ .001 for 9551(□), P ≤ .01 for 9515 (∆), and P ≤ .001 for 9518 (○). Abbreviations: AQP1, aquaglyceroporin; PTR1, pteridine reductase 1; PURO, puromycin acetyltransferase.
Figure 2.
Genomic analysis of Leishmania donovani clinical isolates. A, A phylogenetic network of L. donovani isolates 9515, 9518, and 9551 with parasites of the core population found in the Indian subcontinent (see Supplementary Table 1) [24]. B, Ploidy of the 36 chromosomes of the 3 L. donovani isolates. Abbreviation: ISC, Indian Subcontinent.
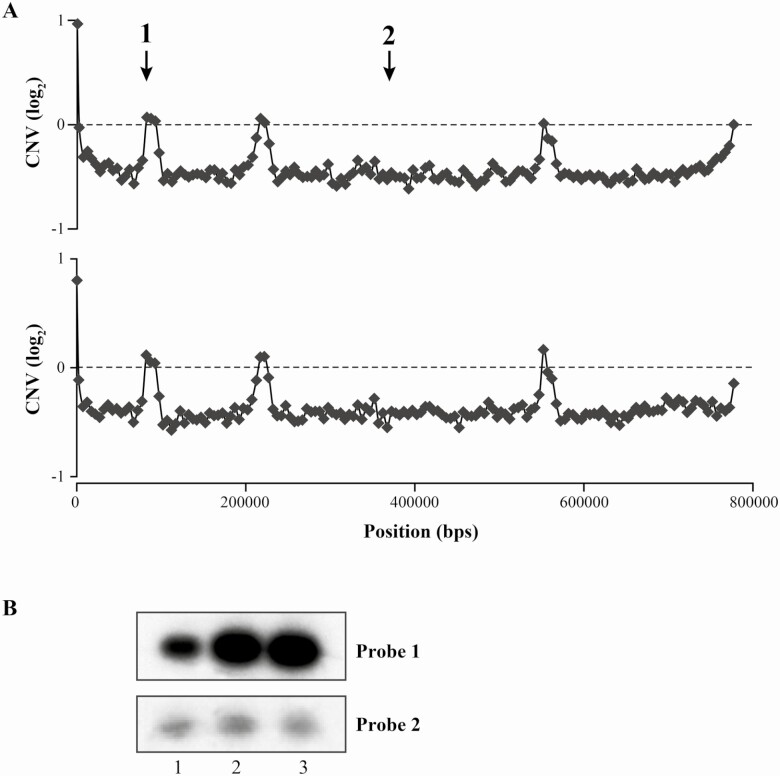
Normalized read depth coverage highlighted potential CNVs of specific genomic loci ranging from 40- to 115-kb amplification or deletion. These are characterized by loci whose normalized read coverage varies in the resistant isolates (9515, 9518) compared with the sensitive one (9551) (Supplementary Figure 1, Figure 3A). Some of these putative CNVs on chromosomes 1, 13, and 23 were shared between the 2 resistant isolates, whereas others on chromosome 7 or 35 were specific to only 1 strain (Supplementary Figure 1). One region amplified on chromosome 23 present that was in the 2 resistant strainscodes for MRPA, a proven marker of resistance to antimony. This CNV, revealed in silico, was confirmed by a Southern blot analysis (Figure 3B). A 700-bp probe within the MRPA coding sequences (probe 1 in Figure 3A) was hybridized to the genomic DNAs of the 3 strains. The hybridization profile confirmed that the copy number of MRPA is amplified by 2-fold in the resistant strains (Figure 3B). A probe hybridizing the LdBPK_231370 gene located outside of the amplified region (probe 2 in Figure 3A) monitored the amount of DNA layered in each lane (Figure 3B).
Figure 3.
Chromosome 23 CNV in Leishmania donovani. A, Log2-transformed 9515/9551 (top) and 9518/9551 (bottom) reads ratio for nonoverlapping 5-kb genomic windows on chromosome 23. The numbers 1 and 2 indicate the position of the probes used for Southern blot hybridization. B, Southern blot of L. donovani DNA digested with PvuII and hybridized with probe 1 (MRPA) and 2 (LdBPK_231370) shown in panel A. Lane 1, L. donovani 9551; lane 2, L. donovani 9515; lane 3, L. donovani 9518. Abbreviations: CNV, copy number variation; MRPA, multidrug resistance protein A.
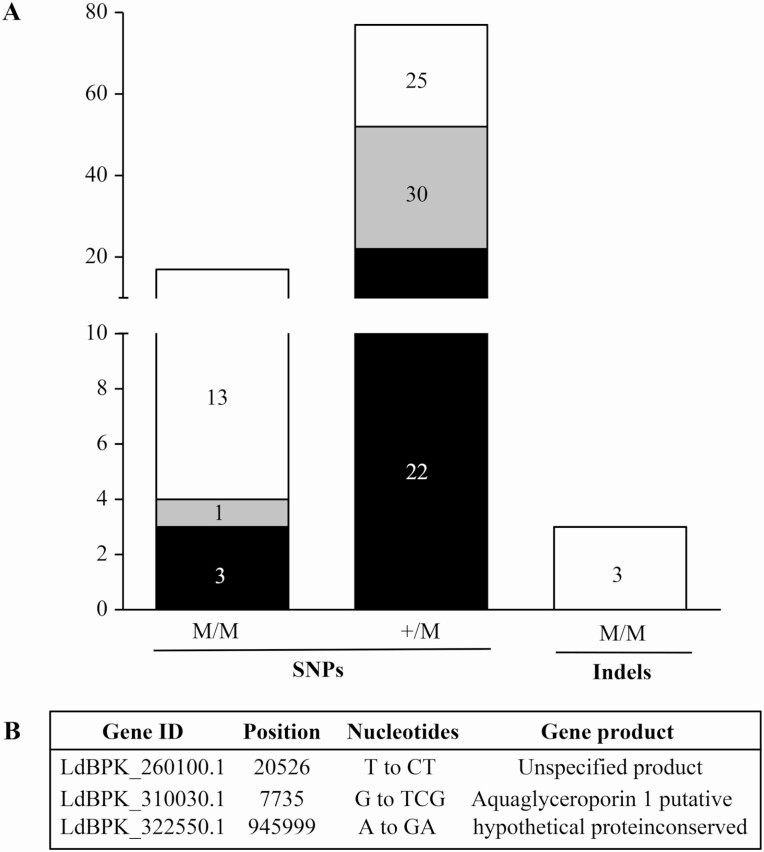
In addition to CNVs we also monitored for SNVs. A total of 97 SNVs (3 indels and 94 single nucleotide polymorphisms [SNPs]) were counted in the coding sequences of both 9515 and 9518 in comparison to 9551 (Figure 4A). Seventy-seven SNPs were heterozygous and 17 were homozygous, 13 of which were shared by both strains (Figure 4A, Supplementary Table 2). The majority of homozygous SNPs were in hypothetical proteins and in kinases. Similarly, the 77 heterozygous SNPs were found predominantly in genes coding for hypothetical proteins or membrane proteins such as amastins and transporters (Supplementary Table 3), but none that would suggest a role in resistance. We could not find mutations in either the coding sequences or intergenic regions of genes coding for trypanothione biosynthesis. Consistent with this lack in mutations, the gamma glutamylcysteine synthetase (GSH1) gene was similarly expressed in the 3 strains (Supplementary Figure 2). The 3 indels were homozygous and shared by both resistant strains (Figure 4A) and one of those homozygous indels consists of a dinucleotide TC insertion in the AQP1 gene at position 7735 of chromosome 31 (Figure 4B, Supplementary Table 3). Chromosome 31 is tetraploid (Figure 2B) and its 4 alleles contained the TC indel in both resistant lines. This insertion in AQP1 was also noted by Imamura et al [24], and was detected in many resistant isolates, including the ISC5 group. This dinucleotide insertion did not modulate the level of AQP1 transcripts (Supplementary Figure 2). This indel is stable as our clinical isolates are routinely passaged in drug-free medium and the indel is maintained.
Figure 4.
Single nucleotide variants in Leishmania donovani antimony-resistant isolates. A, The numbers of SNPs homozygous or heterozygous and of homozygous indels present in 9515 (black), 9518 (gray), or both (white) and absent in 9551 are shown. B, List of the 3 homozygous indels. Abbreviations: SNP, single nucleotide polymorphism; M/M, Homozygous; +/M, Heterozygous.
Genome Editing of AQP1 and the Role of the TC Insertion in Antimony Resistance
While the frameshift insertion in AQP1 and antimony resistance are correlated, experimental validation in the context of clinical isolates is warranted. We first removed by DNA editing the TC insertion in 9515 and 9518 to yield a wild-type sequence of AQP1 in these initially resistant isolates. Second, we introduced the TC dinucleotide in the AQP1 sequence of the initially sensitive 9551 isolate. This was done in strains expressing the Cas9 nuclease. Since those edition events were selected by co-transfection of a PTR1 PURO inactivating cassette, this cassette was introduced as well in all control cells. The integration of the PURO marker in PTR1 in any of the 3 original strains did not change their susceptibility to SbV (Figure 1B). DNA sequencing of the AQP1 gene derived from clones of edited cells confirmed the removal (9515, 9518) or incorporation (9551) of the TC dinucleotide. The antimony susceptibility increased significantly in both strains 9515 and 9518 (unpaired t test, 2-tailed P ≤ .001 and P ≤ .01, respectively) when the TC insertion in AQP1 was removed (Figure 1B). These strains, however, remained slightly more resistant than the 9551 sensitive isolate. When a TC was introduced in the AQP1 gene of 9551, it became significantly resistant to SbV (unpaired t test, 2-tailed P ≤ .001) (Figure 1B). The dinucleotide insertion was also stable since this strain was routinely cultured in the absence of antimonials and the indel and resistance were maintained.
DISCUSSION
This genomic study of L. donovani clinical isolates has highlighted 2 markers of resistance (MRPA and AQP1). The role of MRPA in SbV resistance was proven by episomal transfection first in laboratory-adapted strains [34] and in clinical isolates [10]. Here we proved using DNA editing that AQP1 contributes to SbV resistance in clinical isolates (Figure 1B).
Analysis of in vitro SbIII-resistant mutant has shown that the copy number of AQP1 can be decreased by telomeric gene deletion [28, 35]. Leishmania cells whose AQP1 gene was deleted by standard gene knock-out were also resistant to SbIII [36]. Gene deletion leads to less RNA expression. and this is consistent with studies of cells selected for resistance in vitro [18] or in isolates where reduced expression of AQP1 was associated with SbV resistance in L. donovani [19–21]. Several exceptions were noted, however, and either equal or even increased RNA levels of AQP1 were observed in some L. donovani–resistant isolates [12, 20]. A point mutation in AQP1 was also detected in strains selected for resistance to SbIII in vitro [35]. Resistance may thus arise by the modulation in gene expression (eg, by gene deletion or by SNPs changing RNA stability) or by point mutation in AQP1. Sequencing of clinical isolates [24] including the ones in this study did not support AQP1 gene deletion but instead highlighted a TC insertion in AQP1. This insertion disrupts the AQP1 reading frame and is likely to produce a nonfunctional truncated protein. In this case, there is no need to alter AQP1 RNA levels (Supplementary Figure 2).
By genome editing we proved the role of the AQP1 insertion in resistance to antimony in L. donovani clinical isolates (Figure 1B). The removal of the TC dinucleotide in 9515 or 9518 increased susceptibility, although not to the level of the 9551 sensitive isolate. This suggests that other resistance mechanisms are likely to be present, and it is salient to point out that MRPA was amplified in 9515 and 9518 (Figure 3). Resistance may thus be due to a combination of the indel inactivation of AQP1 and MRPA CNVs. There are additional SNVs (Figure 4, Supplementary Table 2) and CNVs (Figure 2B, Supplementary Figure 1) in the clinical isolates and some may also contribute to resistance or act as compensatory mutations following the inactivation of AQP1. Indeed, AQP1 appears to have multiple physiological roles in solute transport, volume regulation, and osmotaxis [37], and its loss may require compensation. These resistant isolates are fit parasites infecting well macrophages [38] and it can be argued that this fitness may arise by compensatory mutations. While mutations in AQP1 are likely to be the main mechanism for achieving a high level of resistance, as exemplified by the introduction of the TC indel in the sensitive clinical isolate 9551 (Figure 1B), the locus encoding MRPA is often amplified in the main core group of Indian isolates [24]. It has been hypothesized that this amplification constitutes a first line of defense against antimonials before mutations in AQP1 produce more resistance [10]. Interestingly, point mutations [39] or reduced expression [40] of AQP1 or increased expression of MRPA [40] have been described in Iranian cutaneous isolates not responding to therapy. Thus, AQP1 and MRPA may contribute to resistance in other Leishmania species.
Of the 3 markers found in vitro that were also reported to operate in clinical isolates, our study, as others [23, 24], failed to detecting CNVs or SNVs in genes involved in trypanothione biosynthesis. The expression of the gene GSH1, coding for the γ-glutamylcysteine synthetase, the rate-limiting step in glutathione biosynthesis in Leishmania [14], was reported to be altered [9, 12, 17, 19] in resistant isolates. In our limited study, the expression of GSH1 was equal in all parasites independent of their susceptibility status (Supplementary Figure 2).
The antimonials SbV and SbIII accumulate in Leishmania by different transporters [6] and AQP1 only transports the trivalent form [7]. SbV is thought to be reduced to SbIII either in the host macrophage or in the parasite [3]. Since AQP1 only transports SbIII, the results observed here would suggest that reduction takes place predominantly in the macrophage. In summary, we have proven that indels in AQP1 contribute to resistance to SbV in L. donovani clinical isolates.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Hélène Gingras for help with quantitative real-time PCR.
Financial support. This work was supported by the Canadian Institutes of Health Research (grant number FND167283). M. O. is a Canada Research Chair in Antimicrobial Resistance. Infrastructure and equipment were provided by the Canadian Foundation for Innovation. J.-E. P. and M. Q. were supported by the Syndicat des Professeurs et Professeures de l’Université Laval (SPUL) Leadership fellowships.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Alvar J, Vélez ID, Bern C, et al. ; WHO Leishmaniasis Control Team . Leishmaniasis worldwide and global estimates of its incidence. PLoS One 2012; 7:e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lira R, Sundar S, Makharia A, et al. . Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J Infect Dis 1999; 180:564–7. [DOI] [PubMed] [Google Scholar]
- 3. Ponte-Sucre A, Gamarro F, Dujardin JC, et al. . Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Negl Trop Dis 2017; 11:e0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perry M, Wyllie S, Prajapati V, et al. . Arsenic, antimony, and Leishmania: has arsenic contamination of drinking water in India led to treatment- resistant kala-azar? Lancet 2015; 385(Suppl 1):S80. [DOI] [PubMed] [Google Scholar]
- 5. Dey S, Papadopoulou B, Haimeur A, et al. . High level arsenite resistance in Leishmania tarentolae is mediated by an active extrusion system. Mol Biochem Parasitol 1994; 67:49–57. [DOI] [PubMed] [Google Scholar]
- 6. Brochu C, Wang J, Roy G, et al. . Antimony uptake systems in the protozoan parasite Leishmania and accumulation differences in antimony-resistant parasites. Antimicrob Agents Chemother 2003; 47:3073–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gourbal B, Sonuc N, Bhattacharjee H, et al. . Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem 2004; 279:31010–7. [DOI] [PubMed] [Google Scholar]
- 8. Légaré D, Richard D, Mukhopadhyay R, et al. . The Leishmania ATP-binding cassette protein PGPA is an intracellular metal-thiol transporter ATPase. J Biol Chem 2001; 276:26301–7. [DOI] [PubMed] [Google Scholar]
- 9. Mukherjee A, Padmanabhan PK, Singh S, et al. . Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J Antimicrob Chemother 2007; 59:204–11. [DOI] [PubMed] [Google Scholar]
- 10. Dumetz F, Cuypers B, Imamura H, et al. . Molecular preadaptation to antimony resistance in Leishmania donovani on the Indian Subcontinent. mSphere 2018; 3. doi: 10.1128/mSphere.00548-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mittal MK, Rai S, Ashutosh, et al. Characterization of natural antimony resistance in Leishmania donovani isolates. Am J Trop Med Hyg 2007; 76:681–8. [PubMed] [Google Scholar]
- 12. Kumar D, Singh R, Bhandari V, Kulshrestha A, Negi NS, Salotra P. Biomarkers of antimony resistance: need for expression analysis of multiple genes to distinguish resistance phenotype in clinical isolates of Leishmania donovani. Parasitol Res 2012; 111:223–30. [DOI] [PubMed] [Google Scholar]
- 13. Mukhopadhyay R, Dey S, Xu N, et al. . Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci USA 1996; 93:10383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grondin K, Haimeur A, Mukhopadhyay R, Rosen BP, Ouellette M. Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenite-resistant Leishmania tarentolae. EMBO J 1997; 16:3057–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science 1985; 227:1485–7. [DOI] [PubMed] [Google Scholar]
- 16. Mandal G, Wyllie S, Singh N, Sundar S, Fairlamb AH, Chatterjee M. Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology 2007; 134:1679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh N, Chatterjee M, Sundar S. The overexpression of genes of thiol metabolism contribute to drug resistance in clinical isolates of visceral leishmaniasis (kala azar) in India. Parasit Vectors 2014; 7:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol 2005; 57:1690–9. [DOI] [PubMed] [Google Scholar]
- 19. Decuypere S, Rijal S, Yardley V, et al. . Gene expression analysis of the mechanism of natural Sb(V) resistance in Leishmania donovani isolates from Nepal. Antimicrob Agents Chemother 2005; 49:4616–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maharjan M, Singh S, Chatterjee M, Madhubala R. Role of aquaglyceroporin (AQP1) gene and drug uptake in antimony-resistant clinical isolates of Leishmania donovani. Am J Trop Med Hyg 2008; 79:69–75. [PubMed] [Google Scholar]
- 21. Mandal S, Maharjan M, Singh S, Chatterjee M, Madhubala R. Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and -resistant clinical isolates of Leishmania donovani from India. J Antimicrob Chemother 2010; 65:496–507. [DOI] [PubMed] [Google Scholar]
- 22. Leprohon P, Fernandez-Prada C, Gazanion É, Monte-Neto R, Ouellette M. Drug resistance analysis by next generation sequencing in Leishmania. Int J Parasitol Drugs Drug Resist 2015; 5:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Downing T, Imamura H, Decuypere S, et al. . Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 2011; 21:2143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Imamura H, Downing T, Van den Broeck F, et al. . Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. eLife 2016; 5. doi: 10.7554/eLife.12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy G, Dumas C, Sereno D, et al. . Episomal and stable expression of the luciferase reporter gene for quantifying Leishmania spp. infections in macrophages and in animal models. Mol Biochem Parasitol 2000; 110:195–206. [DOI] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKenna A, Hanna M, Banks E, et al. . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukherjee A, Boisvert S, Monte-Neto RL, et al. . Telomeric gene deletion and intrachromosomal amplification in antimony-resistant Leishmania. Mol Microbiol 2013; 88:189–202. [DOI] [PubMed] [Google Scholar]
- 29. Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019; 35:4453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 2006; 23:254–67. [DOI] [PubMed] [Google Scholar]
- 31. Bhattacharya A, Leprohon P, Bigot S, et al. . Coupling chemical mutagenesis to next generation sequencing for the identification of drug resistance mutations in Leishmania. Nat Commun 2019; 10:5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peng D, Tarleton R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genom 2015; 1:e000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gagnon D, Foucher A, Girard I, Ouellette M. Stage specific gene expression and cellular localization of two isoforms of the serine hydroxymethyltransferase in the protozoan parasite Leishmania. Mol Biochem Parasitol 2006; 150:63–71. [DOI] [PubMed] [Google Scholar]
- 34. El Fadili K, Messier N, Leprohon P, et al. . Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob Agents Chemother 2005; 49:1988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monte-Neto R, Laffitte MC, Leprohon P, Reis P, Frézard F, Ouellette M. Intrachromosomal amplification, locus deletion and point mutation in the aquaglyceroporin AQP1 gene in antimony resistant Leishmania (Viannia) guyanensis. PLoS Negl Trop Dis 2015; 9:e0003476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plourde M, Ubeda JM, Mandal G, Monte-Neto RL, Mukhopadhyay R, Ouellette M. Generation of an aquaglyceroporin AQP1 null mutant in Leishmania major. Mol Biochem Parasitol 2015; 201:108–11. [DOI] [PubMed] [Google Scholar]
- 37. Figarella K, Uzcategui NL, Zhou Y, et al. . Biochemical characterization of Leishmania major aquaglyceroporin LmAQP1: possible role in volume regulation and osmotaxis. Mol Microbiol 2007; 65:1006–17. [DOI] [PubMed] [Google Scholar]
- 38. Vanaerschot M, De Doncker S, Rijal S, Maes L, Dujardin JC, Decuypere S. Antimonial resistance in Leishmania donovani is associated with increased in vivo parasite burden. PLoS One 2011; 6:e23120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alijani Y, Hosseini SS, Ahmadian S, et al. . Molecular analysis of aquaglyceroporin 1 gene in non-healing clinical isolates obtained from patients with cutaneous Leishmaniasis from central of Iran. J Arthropod Borne Dis 2019; 13:145–52. [PMC free article] [PubMed] [Google Scholar]
- 40. Mohebali M, Kazemirad E, Hajjaran H, et al. . Gene expression analysis of antimony resistance in Leishmania tropica using quantitative real-time PCR focused on genes involved in trypanothione metabolism and drug transport. Arch Dermatol Res 2019; 311:9–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.