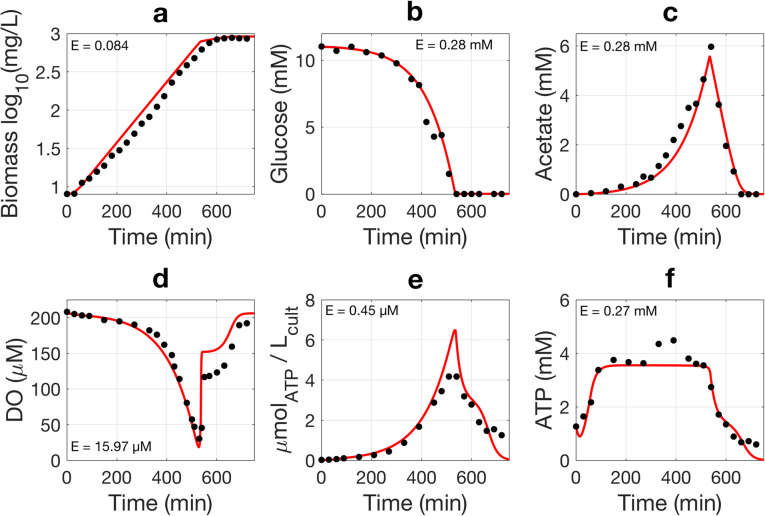
Fig. 6.
Experimental data and model simulation for batch-culture kinetics. The BW25113 strain with the ATP reporter was grown in minimal medium. Black circles represent measured experimental data (means of three independent replicates) and red lines represent model simulation results. Biomass (a) was estimated by multiplying the mass of a single cell with cell counts determined via flow cytometry. Glucose (b), acetate (c), and dissolved oxygen (d) were measured as described in the “Methods” section. ATP was measured by a standard luciferase assay. Population ATP (e) is indicated by the amount of ATP per L of culture. Cellular ATP (f) is calculated from the population ATP, cell counts, and the volume of a single cell. E is the average absolute error of the model in the units of the corresponding y-axis and indicates the model’s goodness of fit for each data fitting. The small E values indicate good fits of our model