Abstract
Introduction
The molecular mechanisms underlying alcoholic liver fibrosis and cirrhosis are not completely understood. Hepatic fibrosis involves the interplay of diverse cells and factors, including hepatic stellate cells (HSCs), Kupffer, NK cells, and T-lymphocyte subsets. Killer-cell immunoglobulin-like receptors (KIR) are membrane receptors involved in mediation between NK and activated HSCs, regulating NK cell function through their interaction with HLA-I molecules. The aim of this study was to analyse the genetic association between KIR genes and the susceptibility to or protection from alcoholic cirrhosis (AC) in a cohort of male AC patients undergoing liver transplantation (LT) with and without concomitant viral infections.
Material and methods
KIR genotyping was performed in nuclear DNA extracted from 281 AC patients and compared with 319 male controls.
Results
Significant differences between total AC patients and healthy controls were only found in the case of KIR2DL2 and KIR2DS5. KIR2DL2 was significantly underrepresented in non-viral AC patients (52.6% vs. 63.3%; p = 0.015), while patients heterozygous for KIR2DL2 were also underrepresented in the non-viral AC group compared with controls (p = 0.034). KIR2DS5 was overrepresented in this group compared with healthy controls (p = 0.002). All these observations were only evident in AC patients older than 54 years old.
Conclusions
Our data suggest a contrary effect of KIR2DL2 and KIR2DS5 in AC patients older than 54 years, in whom the presence of KIR2DL2 appears to be protective against AC, whereas the presence of KIR2DS5 seems to promote the fibrotic process, particularly in patients with no associated viral infection.
Keywords: alcohol, cirrhosis, human clinical toxicology, KIr genes, NK cells
Introduction
Alcoholism is a major socio-economic and medical problem worldwide, and a major risk factor for chronic liver disease, which includes fatty liver, alcoholic hepatitis, fibrosis, cirrhosis and hepatocellular carcinoma [1–6]. Alcoholic cirrhosis (AC) is the most common indication for liver transplantation (LT), which accounts for approximately 33% of all primary transplants in Europe [7–11].
However, the molecular and cellular mechanisms underlying alcoholic liver fibrosis and its end-stage consequence, cirrhosis, are not completely understood [12–15], although it is generally believed that many factors and different types of cells interact in their development [16, 17].
The liver is considered an organ with a predominantly innate immunity, and rich in Kupffer and NK cells, as well as in T-lymphocyte subsets [18–20]. There is evidence that innate immunity is involved in controlling the development and progression of liver fibrosis [21–23], and that liver hepatic stellate cells (HSCs) are activated as a consequence of chronic liver injury, playing a central role in the production of extracellular matrix proteins (ECM), whose accumulation induces fibrosis [24, 25]. The HSC activation appears to be controlled by many types of cytokines, growth factors, immune cells (especially NK and T-lymphocyte subsets) and other factors, including alcoholic detoxification products such as acetaldehyde [23–26].
Recent studies on killer-cell immunoglobulin-like receptors (KIR) and HLA ligands suggested that they play a role in liver immunopathology [27–29]. The NK cell function is regulated through a balance of specific interactions between various membrane receptors, including KIRs, and human leukocyte antigen IDEM (HLA) class I molecules [30].
KIR receptors are encoded by a family of genes located in the leucocyte receptor complex (LRC) of chromosome 19q13.4 [28, 31, 32], which exhibit a high degree of polymorphism and differ among individuals [33, 34]. According to the cytoplasmic tail structure, two kinds of functional KIR can be distinguished – inhibitory KIRs (iKIRs), with a long cytoplasmic tail with at least one immunoreceptor tyrosine-based inhibitory motif (ITIM), and activating KIRs (aKIR), with a short cytoplasmic tail in which ITIM motifs are absent [35, 36]. Thus, there are six iKIR (KIR3DL1-3, KIR2DL1-3), six aKIR (KIR3DS1 and KIR2DS1-5) genes, and one KIR2DL4 gene which encodes a receptor with both inhibitory and activating functions.
The aim of the present study was to analyse the influence of KIR genes on the susceptibility to or protection against AC in a cohort of male patients undergoing LT with and without concomitant viral infections, in an attempt to identify biomarkers of susceptibility to AC in order to design new strategies to prevent, detect and reduce AC in men with high alcohol consumption.
Material and methods
Studied populations
A cohort of 281 male AC patients undergoing LT, recruited between 1990 and 2013, was analysed retrospectively. The clinical and biochemical characteristics and short- and long-term survival of the same cohort of AC patients have been published previously [9, 11]. In this study, the AC patients were divided into two age groups and receiver operation characteristic (ROC) curves were used to find an optimal cut-off value categorizing it into young and old groups. A range of optimal values was obtained between 51.5 and 55.5 years, with sensitivity of 0.700–0.573, and specificity of 0.691–0.765 respectively. The final cut-off value of 54 years was chosen, making it correspond to the sample median value, which corresponds to the clinical diagnosis of the disease. A total of 143 AC patients were 54 years old or less and the remaining 138 AC patients were older than 54 years old.
Furthermore, a series of 319 male individuals matched in age was used as a healthy control cohort to compare with the total male AC population. In all cases, both populations were compared between similar median ages.
Written informed consent was obtained from all patients and controls in this study. The study protocol was approved by the institutional ethics committee study in accordance with the Helsinki Declaration of 2000.
Alcoholic cirrhosis diagnosis
Alcoholic cirrhosis was diagnosed by reference to clinical, radiological, and biochemical parameters [37]. In cases of negative self-reporting concerning alcohol consumption, the opinion of relatives was taken into consideration. Data on alcoholic beverage type and average intake could not be obtained. In most cases, there were no symptoms of cirrhosis in the first stage of the disease, so the diagnosis was made after a scan, ultrasound or clinical examination. In other cases, the disease remained undetected until the second stage of decompensated cirrhosis, when complications such as ascites, upper gastrointestinal bleeding and encephalopathy appeared. Cases of suspected cirrhosis were confirmed using specific analysis and imaging technologies. It is important to note that the degree of hepatic fibrosis of all patients included in this study was grade F4 (METAVIR score), at the time of inclusion on the waiting list for LT.
Viral infection diagnosis
Hepatitis C virus (HCV) pre-infection was determined using a qualitative immunoassay (AxSYM HCV v3.0; Abbott, Wiesbaden-Delkenheim, Germany) to detect the presence of anti-HCV antibodies, and the results were confirmed by immunoblotting technology (recombinant immunoblot assay) or reverse transcription and polymerase chain reaction (PCR) (REAL; Durviz, Valencia, Spain), following the manufacturer’s indications. Hepatitis B viral infection (HBV) was determined by measuring the HBV surface antigen using a radioimmunological method (Sorin Biomedica, Perugia, Italy). To determine cytomegalovirus (CMV) infection, anti-CMV-IgG antibodies were tested by immunoassay (Liason CMV-IgG; DiaSorin, Saluggia, Italy); CMV infection was diagnosed taking into account an IgG antibody level of ≥ 0.6 IU/ml. Data of these positive cases were confirmed using a real-time PCR (LightCycler CMV-Quant-kit; Roche, Indianapolis, IN). According to the presence or not of virus infections, male AC patients were classified into viral (n = 68) and non-viral AC patient groups (n = 213).
Sample collection and DNA preparation
Samples obtained from peripheral blood as sources of DNA were used for this genetic association study. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany), as recommended by the manufacturer.
KIR genotyping
Genotyping of the KIR genes was performed using the multiplex KIR-SSO typing kit from Tepnel Lifecodes Corporation (Ref: 545110, Connecticut, USA). This product consists of a mixture of locus-specific oligonucleotide probes coupled to colour-coded microspheres (Luminex Corp) and two PCR reactions for the amplification of KIR-exons 4, 5, 7, 8, and 9. To type each sample, PCR was performed and the product was hybridized with the SSO-probe mixture using the manufacturer’s protocol. After hybridization, the sample plate was placed in a Luminex instrument for analysis [38–40]. In addition, KIR genotypes were confirmed by PCR-SSP [41]. This method identifies inhibitory KIR2DL1-3, KIR3DL1-3, activating KIR2DS1-S5 and KIR3DS1 genes, as well as KIR2DL4. The genes KIR2DL5A and KIR2DL5B could not be distinguished, while KIR2DP1 and KIR3DP1 pseudogenes were identified. Given that KIR2DL2 is generally closely associated with KIR2DS2 and that their individual effects could not be distinguished in our population, both genes were studied together.
For further analysis, KIR genotype profiles were classified in accordance with the Allele Frequencies KIR Database website [42] and each genotype was named according to the genotype number (ID) defined in this website.
KIR haplotypes A and B and genotype assignment
The KIR gene content was used to infer the A and B KIR haplotypes and to assign three genotypes, AA, BB and AB, to individuals [42]. Individuals carrying only KIR3DL3, 2DL3, 2DL1, 2DP1, 3DP1, 2DL4, 3DL1, 2DS4, 3DL2 genes of the A KIR haplotypes were considered to be homozygous for the A haplotype, and therefore as having the KIR genotype AA [42]. Conversely, those individuals who lacked one or more of the KIR2DL1, 2DL3, 3DL1 or 2DS4 genes were regarded as carrying two copies of the B KIR haplotype (BB genotype).
The remaining individuals were considered to be AB heterozygous or bearing AB genotypes. These individuals possessed all nine genes present in the A haplotype, as well as one or more of the B haplotype specific genes (KIR2DL2, 2DL5, 2DS1, 2DS2, 2DS3, 2DS5 and 3DS1). AB and BB genotypes were also referred to together as KIR genotype BX [43], and each individual KIR gene was also assigned to the centromeric or telomeric regions in the KIR gene complex [44, 45].
Statistical analysis
Demographic data and results of the KIR gene analysis were collected in a database (Microsoft Access 11.0; Microsoft corporation, Seattle, WA), and statistical analysis was performed using the SPSS 15.0 software (SPSS Inc., Chicago IL, USA). To detect differences in the frequencies of KIR genes and genotypes, Pearson’s χ2 and two-tailed Fisher’s exact tests were run to compare categorized variables between groups, and a two-sided Student T test and the nonparametric Mann-Whitney test were used to compare mean values. For continuous variables such as age and AC, a receiver operation characteristic (ROC) curve was used to acquire a cut-off value for stratification. Multivariable logistic regression analysis was applied to confirm positive associations. A level of p < 0.05 was accepted as statistically significant. Odds ratios (OR) and their 95% confidence intervals (CI) were calculated to estimate relative risk.
Results
Clinical characteristics of patients
Analysis of the different types of viral infections revealed that most of the AC patients (75.8%; n = 213) did not present any kind of viral infection. Thus, viral infected AC patients represented 23.4% (n = 68) of the total number of AC patients, with 61.8% of them infected by HCV, 20.6% by HBV, and 5.9% by CMV. Additionally, 5.9% of the AC patients presented HCV associated with HBV infection, 2.9% HCV associated with CMV, and 2.9% HBV combined with CMV.
KIR2DL2 and KIR2DL3 zygosity was differently associated with viral and non-viral AC patients
The frequency of five iKIRs was determined in AC patients and healthy controls (Table I, top). Univariate analysis revealed no significant differences in the iKIR gene frequency between AC patients and healthy controls, except in the case of KIR2DL2+ patients, who were significantly underrepresented within the total group of AC patients (53%; p = 0.013). In addition, a statistically significant reduced frequency of KIR2DL2 was observed when the non-viral AC group was compared with the control group (52.6% vs. 63.3%; p = 0.015).
Table I.
KIR gene frequencies in male patients with alcoholic cirrhosis and healthy controls
| KIR genes* | P/A | Healthy controls (N = 319) n (%) | Alcoholiccirrhosis (AC) | P2 | P3 | P4 | |||
|---|---|---|---|---|---|---|---|---|---|
| Total AC patients (N = 281) n (%) | P1 | Non-viral (N = 213) n (%) | Viral (N = 68) n (%) | ||||||
| iKIRs: | |||||||||
| 2DL1 (S1-) | + | 197 (61.8) | 160 (56.9) | 0.244 | 115 (54.0) | 45 (66.2) | 0.088 | 0.581 | 0.092 |
| – | 122 (38.2) | 121 (43.1) | 98 (46.0) | 23 (33.8) | |||||
| 2DL2 | + | 202 (63.3) | 149 (53.0) | 0.013a | 112 (52.6) | 37 (54.4) | 0.015d | 0.173 | 0.889 |
| – | 117 (36.7) | 132 (47) | 101 (47.4) | 31 (45.6) | |||||
| 2DL3 | + | 279 (87.5) | 249 (88.6) | 0.707 | 190 (89.2) | 59 (86.8) | 0.586 | 0.842 | 0.661 |
| – | 40 (12.5) | 32 (11.4) | 23 (10.8) | 9 (13.2) | |||||
| 3DL1 | + | 304 (95.3) | 268 (95.4) | 1.000 | 201 (94.4) | 67 (98.5) | 0.689 | 0.325 | 0.200 |
| – | 15 (4.7) | 13 (4.6) | 12 (5.6) | 1 (1.5) | |||||
| 2DL5 | + | 170 (53.3) | 158 (56.2) | 0.511 | 121 (56.8) | 37 (54.4) | 0.477 | 0.894 | 0.780 |
| – | 149 (46.7) | 123 (43.8) | 92 (43.2) | 31 (45.6) | |||||
| aKIRs: | |||||||||
| 2DS1 (L1+) | + | 119 (37.3) | 119 (42.3) | 0.211 | 96 (45.1) | 23 (33.8) | 0.087 | 0.678 | 0.121 |
| – | 200 (62.7) | 162 (57.7) | 117 (54.9) | 45 (66.2) | |||||
| 2DS2 (L2+) | + | 201 (63.0) | 146 (52) | 0.006b | 109 (51.2) | 37 (54.4) | 0.007e | 0.217 | 0.677 |
| – | 118 (37.0) | 135 (48) | 104 (48.8) | 31 (45.6) | |||||
| 2DS3 | + | 107 (33.5) | 93 (33.1) | 0.931 | 65 (30.5) | 28 (41.2) | 0.508 | 0.263 | 0.107 |
| – | 212 (66.5) | 188 (66.9) | 148 (69.5) | 40 (58.8) | |||||
| 2DS4 | + | 305 (95.6) | 266 (94.7) | 0.704 | 199 (93.4) | 67 (98.5) | 0.323 | 0.486 | 0.128 |
| – | 14 (4.4) | 15 (5.3) | 14 (6.6) | 1 (1.5) | |||||
| 2DS5 | + | 86 (27.0) | 99 (35.2) | 0.033c | 85 (39.9) | 14 (20.6) | 0.002f | 0.360 | 0.004g |
| – | 233 (73.0) | 182 (64.8) | 128 (60.1) | 54 (79.4) | |||||
| 3DS1 | + | 129 (40.4) | 132 (47.0) | 0.117 | 105 (49.3) | 27 (39.7) | 0.050h | 1.000 | 0.209 |
| – | 190 (59.6) | 149 (53.0) | 108 (50.7) | 41 (60.3) | |||||
+ presence; – absence; N – total number of individuals; n – number of individuals with the presence or absence of KIR gene. OR – odds ratio with a confidence interval (CI) of 95%. Comparisons were made by the two-sided Fisher´s exact test. Framework genes and pseudogenes were not included. P-values marked in bold are statistically significant (p < 0.05). P1 – p-value obtained comparing total AC patients versus healthy donors; P2 and P3 – p-values obtained comparing non-viral AC patients and viral AC patients versus healthy controls, respectively; P4 – p-value obtained comparing non-viral versus viral AC patients. P5 – p-value obtained comparing VHC cirrhosis patients versus healthy controls.
OR = 1.530; 95% CI: 1.103–2.120, p = 0.013;
OR = 1.575; 95% CI: 1.137–2.183, p = 0.006;
OR = 0.679; 95% CI: 0.479–0.961, p = 0.033;
OR = 1.557; 95% CI: 1.095–2.215, p = 0.015;
OR = 1.625; 95% CI: 1.143–2.311, p = 0.007;
OR = 0.556; 95% CI: 0.384–0.804, p = 0.002;
OR = 2.561; 95% CI: 1.339–4.900, p = 0.004;
OR = 0.698; 95% CI: 0.492–0.991, p = 0.050.
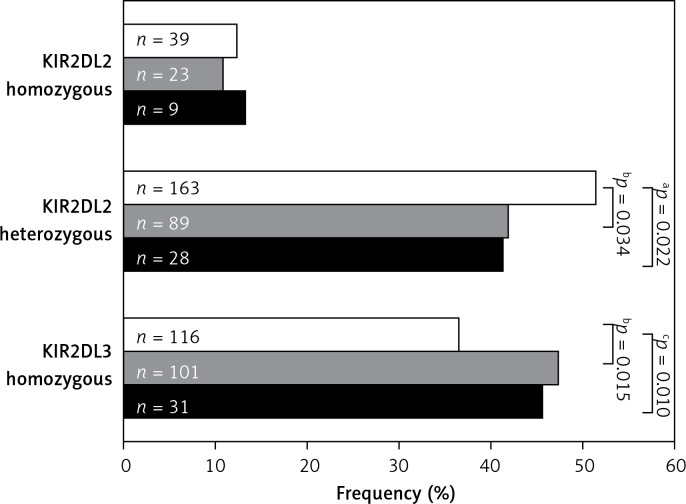
KIR2DL2 and KIR2DL3 homozygosity was also analysed in AC patients and healthy controls (Figure 1). This analysis revealed that KIR2DL2 (L2+/L2+) homozygosis was similarly distributed in controls and total AC patients (12.3% vs. 11.4%; p = 0.801), while heterozygous KIR2DL2 (L2+/L3+) combinations were underrepresented in total AC patients with respect to the controls (41.6% vs. 51.3%; p = 0.022). On the other hand, differences between AC groups and controls were only statistically significant when non-viral AC patients were compared with the controls (41.8% vs. 51.3%; p = 0.034). As regard homozygosis in KIR2DL3 (L3+/L3+), a significant increase was observed in total AC patients compared with the healthy controls (47% vs. 36.5%; p = 0.010), but when viral and non-viral groups of patients were compared with the controls, significant differences were only found in the non-viral AC group (47.4% vs. 36.5%; p = 0.015).
Figure 1.
Analysis of KIR2DL2/S2 and KIR2DL3 zygosity in AC patients and healthy controls. White bars designate healthy control population, grey bars designate non-viral AC patients and solid bars represent viral AC patients. KIR – killer-cell immunoglobulin-like receptors. The p-value was determined by two-sided Fisher’s exact test: aOR = 1.474; 95% CI: 1.067–2.037, p = 0.022; bOR = 0.683; 95% CI: 0.481–0.969, p = 0.034; cOR = 0.648; 95% CI: 0.467–0.899, p = 0.010; dOR = 1.557; 95% CI: 1.095–2.215, p = 0.015
It should be noted that the same effects on these iKIR gene combinations was observed in AC patients older than 54 years, but not in those patients of the AC group with a median age lower than or equal to 54 years (Table II).
Table II.
Analysis of KIR2DL2/S2+ and KIR2DS5+ frequencies in male AC patients and healthy controls according to median age of population
| KIR genes | Median age | Healthy controls n (%) | Alcoholic cirrhosis (AC) | P2 | P3 | P4 | |||
|---|---|---|---|---|---|---|---|---|---|
| Total AC patients n (%) | P1 | Non-viral n (%) | Viral n (%) | ||||||
| 2DL2 + | Total | 202 (63.3) | 149 (53.0) | 0.013a | 112 (52.6) | 37 (54.4) | 0.015b | 0.173 | 0.889 |
| < 54 years | 100 (60.2) | 77 (53.8) | 0.299 | 51 (56.0) | 26 (50.0) | 0.596 | 0.202 | 0.492 | |
| > 54 years | 102 (66.7) | 72 (52.2) | 0.012c | 61 (50) | 11 (68.8) | 0.007d | 1.000 | 0.190 | |
| 2DS2 (L2+) | Total | 201 (63.0) | 146 (52) | 0.006e | 109 (51.2) | 37 (54.4) | 0.007f | 0.217 | 0.677 |
| < 54 years | 99 (59.6) | 77 (53.8) | 0.357 | 51 (56.0) | 26 (50.0) | 0.598 | 0.261 | 0.492 | |
| > 54 years | 102 (66.7) | 69 (50) | 0.004g | 58 (47.5) | 11 (68.8) | 0.002h | 1.000 | 0.182 | |
| 2DS5 + | Total | 86 (27.0) | 99 (35.2) | 0.033i | 85 (39.9) | 14 (20.6) | 0.002j | 0.360 | 0.004k |
| < 54 years | 45 (27.1) | 44 (30.8) | 0.529 | 33 (36.3) | 11 (21.2) | 0.156 | 0.469 | 0.063 | |
| > 54 years | 41 (26.8) | 55 (39.9) | 0.024l | 52 (42.6) | 3 (18.8) | 0.007m | 0.765 | 0.102 | |
N – total number of individuals; n – number of individuals with the presence or absence of KIR gene. OR – odds ratio with a confidence interval (CI) of 95%. Comparisons were made by the two-sided Fisher’s exact test. Framework genes and pseudogenes were not included. P-values marked in bold are statistically significant (p < 0.05). P1 – p-value obtained comparing total AC patients versus healthy donors; P2 and P3 – p-values obtained comparing non-viral AC patients and viral AC patients versus healthy controls, respectively; P4 – p-value obtained comparing non-viral versus viral AC patients.
OR = 1.530; 95% CI: 1.103–2.120, p = 0.013;
OR = 1.557; 95% CI: 1.095–2.215, p = 0.015;
OR = 1.833; 95% CI: 1.141–2.945, p = 0.012;
OR = 2.000; 95% CI: 1.227–3.261, p = 0.007;
OR = 1.575; 95% CI: 1.137–2.183, p = 0.006;
OR = 1.625; 95% CI: 1.143–2.311, p = 0.007;
OR = 2.561; 95% CI: 1.339–4.900, p = 0.004;
OR = 0.556; 95% CI: 0.384–0.804, p = 0.002;
OR = 0.679; 95% CI: 0.479–0.961, p = 0.033;
OR = 2.207; 95% CI: 1.353–3.599, p = 0.002;
OR = 2.000; 95% CI: 1.245–3.212, p = 0.004;
OR = 2.000; 95% CI: 1.245–3.212, p = 0.024;
OR = 1.698; 95% CI: 1.492–2.291, p = 0.007.
Activating KIR genes are more abundant in AC patients
The frequency of the six aKIR genes was determined in the total and both AC groups of patients and in controls (Table I, bottom). The univariate analysis showed a statistically significant decrease in KIR2DS2 frequency in total AC patients compared with the healthy controls (52% vs. 63%, respectively; p = 0.006). This lower frequency of KIR2DS2 observed in patients was only statistically significant in the non-viral group of AC patients compared to controls (51.2% vs. 63%; p = 0.007).
In contrast, KIR2DS5 was overrepresented in total AC patients with respect to the healthy controls (35.2% vs. 27%; p = 0.033). This higher frequency of KIR2DS5 with respect to the controls was also significant in the non-viral AC patients (39.9% vs. 27%; p = 0.002) but not in those AC patients affected by viral infections (20.6% vs. 27%; p = 0.360). In addition, the frequency of KIR2DS5 was significantly higher in the non-viral AC group than in the viral AC patients (39.9% vs. 20.6%; p = 0.004).
Both KIR2DS1 and KIR3DS1 genes also showed a higher frequency in the non-viral AC patients with respect to healthy controls, but only in the case of KIR3DS1 could the difference be considered of borderline statistical significance (49.3% vs. 40.4%; p = 0.050).
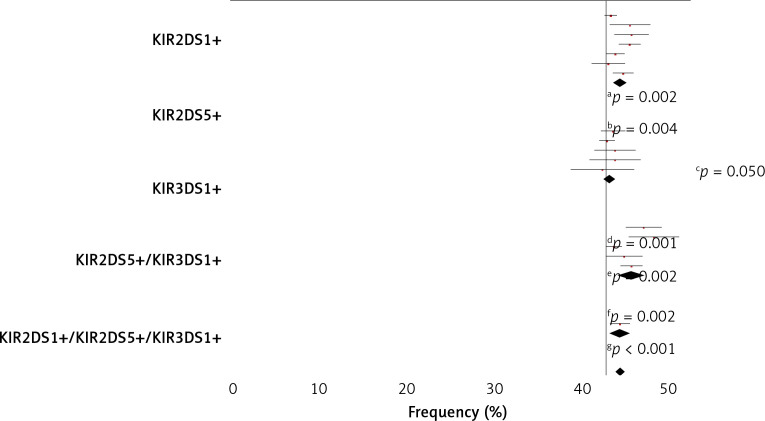
Subsequently, the frequency of KIR2DS1+ plus KIR3DS1+ in association with KIR2DS5+ in both AC patients and control was also analysed (Figure 2). In all cases, a higher frequency of these three associated genes was observed in the non-viral patients than in the viral AC patients, but in no case were these three aKIR genes seen to operate synergically.
Figure 2.
Activating KIR gene frequency in AC patients and healthy controls. Telomeric activating KIR genes, KIR2DS1, KIR2DS5, KIR3DS1 and their combinations, were considered. White bars designate healthy control population; grey bars alcoholic cirrhotic liver patients without viral infection and solid bars alcoholic cirrhotic liver patients with viral infection. KIR – killer-cell immunoglobulin like receptors. P-value was determined by two-sided Fisher’s exact test: aOR = 1.582; 95% CI: 1.151–2.173, p = 0.006; bOR = 2.709; 95% CI: 1.429–5.138, p = 0.002; cOR = 1.434, 95% CI: 1.057–1.946; dOR = 0.595; 95% CI: 0.432–0.819; eOR = 0.350; 95% CI: 0.181–0.674; p = 0.001
It should be noted that the effects observed for iKIR were also observed in AC patients older than 54 years but not in those AC patients less than or equal to 54 years (Table III). Finally, logistic regression multivariable analysis was performed in order to confirm the independent effects of KIR2DS5 (p = 0.021) and KIR2DL2 (p = 0.009) (Table IV).
Table III.
Analysis of KIR2DL2/S2 and KIR2DL3 zygosity in AC patients classified according to median age of population and healthy controls
| KIR genes | Median age | Healthy controls n (%) | Alcoholic cirrhosis (AC) | P2 | P3 | P4 | |||
|---|---|---|---|---|---|---|---|---|---|
| Total AC patients n (%) | P1 | Non-viral n (%) | Viraln (%) | ||||||
| KIR2DL2+ homozygous | Total | 39 (12.3) | 32 (11.4) | 0.801 | 23 (10.8) | 9 (13.2) | 0.680 | 0.840 | 0.661 |
| < 54 years | 20 (12.0) | 18 (12.6) | 1.000 | 12 (13.2) | 6 (11.5) | 0.844 | 1.000 | 1.000 | |
| > 54 years | 19 (12.4) | 14 (10.1) | 0.583 | 11 (9.0) | 3 (18.8) | 0.438 | 0.442 | 0.208 | |
| KIR2DL2+/L3+ heterozygous | Total | 163 (51.3) | 117 (41.6) | 0.022a | 89 (41.8) | 28 (41.2) | 0.034b | 0.143 | 1.000 |
| < 54 years | 80 (48.2) | 59 (41.2) | 0.252 | 39 (42.8) | 20 (38.5) | 0.435 | 0.265 | 0.724 | |
| > 54 years | 83 (54.2) | 58 (42.0) | 0.046c | 50 (41.0) | 8 (50) | 0.030d | 0.796 | 0.593 | |
| KIR2DL3+ homozygous | Total | 116 (36.5) | 132 (47.0) | 0.010e | 101 (47.4) | 31 (45.6) | 0.015f | 0.171 | 0.889 |
| < 54 years | 65 (39.2) | 66 (46.2) | 0.248 | 40 (43.9) | 26 (50) | 0.508 | 0.198 | 0.492 | |
| > 54 years | 51 (33.3) | 66 (47.8) | 0.012g | 61 (50) | 5 (31.3) | 0.007h | 1.000 | 0.190 | |
N – total number of individuals; n – number of individuals with the presence or absence of KIR gene. OR – odds ratio with confidence interval (CI) of 95%. Comparisons were made by two-sided Fisher’s exact test. Framework genes and pseudogenes were not included. P-values marked in bold are statistically significant (p < 0.05). P1 – p-value obtained comparing total AC patients versus healthy donors; P2 and P3 – p-values obtained comparing non-viral AC patients and viral AC patients versus healthy controls, respectively; P4 – p-value obtained comparing non-viral versus viral AC patients.
OR = 1.474: 95% CI: 1.067–2.037, p = 0.022;
OR = 1.465; 95% CI: 1.032–2.079, p = 0.034;
OR = 1.635; 95% CI: 1.028–2.601, p = 0.046;
OR = 1.707; 95% CI: 1.055–2.762, p = 0.030;
OR = 0.648; 95% CI: 0.467–0.899, p = 0.010;
OR = 0.637; 95% CI: 0.447–0.906, p = 0.015;
OR = 0.545; 95% CI: 0.340–0.876, p = 0.012;
OR = 0.500; 95% CI: 0.307–0.815, p = 0.007.
Table IV.
Logistic regression multivariable analysis for KIR in healthy controls and alcoholic cirrhotic patients
| Group of patients | OR (95% CI) | P-value |
|---|---|---|
| Age | 1.017 (1.004–1.029) | 0.009 |
| KIR2DL1 | 0.622 (0.128–3.008) | 0.554 |
| KIR2DL2/S2 | 0.565 (0.369–0.866) | 0.009 |
| KIR2DL3 | 0.878 (0.461–1.671) | 0.692 |
| KIR2DL5 | 0.730 (0.356–1.498) | 0.391 |
| KIR2DS1 | 0.755 (0.315–1.806) | 0.527 |
| KIR2DS3 | 1.362 (0.770–2.408) | 0.288 |
| KIR2DS4 | 0.850 (0.373–1.937) | 0.699 |
| KIR2DS5 | 2.287 (1.135–4.608) | 0.021 |
| KIR3DL1/S1 | 1.296 (0.549–3.062) | 0.554 |
In all cases, comparisons were made between each group of patients and the control group as the reference group. OR, odds ratio with a confidence interval (CI) of 95%. Framework genes and pseudogenes were not included. P-values marked in bold are statistically significant (p < 0.05).
KIR genotype profiles in alcoholic cirrhosis patients
KIR genotype profiles in total AC patients and the viral and non-viral subgroups are summarized in Figure 3. All the above-mentioned genotypes are referred to in the Allele Frequencies KIR Database.
Figure 3.
KIR genotype profiles in total alcoholic cirrhosis patients and in those with and without viral infection. The ID genotypes refer to the genotype classification according to the allele frequencies website (http://www.allelefrequencies.net). The figure represents the frequency of each genotype and indicates the number of individuals carrying a particular genotype in each group. Genotype groups are defined in Material and methods. Each genotype is classified taking into account the total number of KIR genes as well as inhibitory and activating KIR genes. KIR2DL5 was designated as a centromeric KIR gene. Dark boxes represent the KIR genes present and white boxes represent the absent genes in tested individuals
n – number of each genotype, KIR – killer-cell immunoglobulin-like receptors, iKIR – inhibitory killercell immunoglobulin-like receptors, aKIR – activatory killer-cell immunoglobulin-like receptors.
Thirty-nine different KIR genotypes were identified in total AC patients, 38 of which (70.8%) were present as part of the BX genotype, while only 29.2% were integrated in the AA genotype. Similar frequencies of AA and BX genotypes were found in the healthy controls (data not shown).
The most frequent genotypes detected in viral and non-viral AC patients were ID1 (AA) and ID4 (BX), while the ID2 and the ID3 genotypes were less frequent in viral AC patients than in the non-viral AC group. The remaining genotypes had a low representation in both groups of patients. Lastly, the possible influence of the number of total KIR genes, and total aKIR and iKIR genes on AC was also analysed, but no difference between AC patients and healthy controls was observed (data not shown).
Discussion
The current work presents the first retrospective study of the KIR gene repertoire (aKIRs and iKIRs) to be performed in AC male patients undergoing LT, with or without concomitant viral infections. This genetic association study revealed differences in KIR gene distribution when total AC or non-viral and viral AC groups were compared with the controls, suggesting an opposite effect of KIR2DL2 and KIR2DS5 genes in AC patients older than 54 years.
It is well established that chronic alcohol drinking is the main cause of liver fibrosis, which is accelerated in patients with viral infection [9, 11, 13, 14]. After chronic liver injury, hepatocyte damage and activated Kupffer cells lead to the activation of hepatic stellate cells (HSC), producing a large amount of ECM, which are accumulated within the subendothelial space of Disse, and generate liver fibrosis [26, 46].
In the present study, male AC patients who underwent LT had a mean age of 54 years. Similar data were observed in other studies on different populations and periods of time [47–49]. It should be noted that slight differences with respect to this mean age have been described in other studies, suggesting a significant correlation between ethnicity and age of cirrhosis diagnosis, and the possibility of an ethnic and/or genetic basis for cirrhosis [50].
There is wide evidence suggesting that innate immunity is involved in controlling liver fibrosis [26, 51, 52]. More specifically, liver NK cells may be engaged in the inhibition of liver fibrosis and liver regeneration [26, 51, 53].
Under normal conditions and in the absence of fibrotic disease, NK cells are inhibited through interaction of their iKIR and membrane MHC-I molecules of the silent HSCs, which trigger the concomitant inhibition of HSC [51]. However, such inhibition may itself be inhibited as a result of abusive alcohol consumption, which down-regulates class I MHC expression and leads to over-expression of RAE-1 (retinoic acid early inducible-1), thus activating HSCs and making them susceptible to NK killing [26].
In this sense, in vitro studies have demonstrated the beneficial antifibrotic effect of NK cells in incipient liver fibrosis at the onset of liver disease characterized by the presence of early activated hepatic stellate cells (HCSs). Hence, NK cells were shown to inhibit or ameliorate liver fibrosis via the direct killing of early activated HSCs, through the production of antifibrotic substances such as IFN-γ, or HSC proliferation induced by interactions between NK and HSC that are dependent on KIR, NKG2D or granzyme [21, 51, 53].
Nonetheless, interactions between NK and HSC cells have not previously been studied in the final stages of AC, when HSCs are totally activated and there is no possibility of hepatic regeneration. Whatever the case, the participation of iKIR receptors has long been accepted, while the participation of activating aKIR receptors remains to be studied.
To increase our knowledge in this respect, the current study reports differences in the KIR gene repertoire (aKIRs and iKIRs) of healthy individuals and AC patients. When AC patients and controls were compared, a significant decrease in the KIR2DL2/S2 frequency was seen in the non-viral group of AC patients. This result suggests that the presence of KIR2DL2/S2 protects against AC disease development in individuals free of viral infections, while the same kind of AC disease protection could not be confirmed in the viral group AC patients, although they showed similar trends.
Unfortunately, this supposed protection could not be attributed to either KIR2DL2 or KIR2DS2 genes, since these encoding receptors showed contrary inhibitory and activating functions in NK cells, and, particularly, because they showed strong linkage disequilibrium. This means that it is impossible to know whether the observed beneficial effect in preventing AC corresponds to a particular KIR2D gene or involves both genes. A possible explanation for this observed protective effect in non-viral AC patients could be the lower representation of the stronger inhibitory KIR2DL2 gene, which could, in turn, prevent NK cell inhibition, and indirectly favour an increase in activating NK cells, establishing signals to kill active HSCs and so diminish any fibrotic effects [54].
Recently, Motavaf et al. [55] were be able to amplify the anti-fibrotic activity of NK cells and ameliorate hepatic fibrosis in mouse and human co-cultures by iKIR silencing of specific siRNA in cells from subjects with advanced liver disease. This information agrees with our observations about the protective effect of KIR2DL2 against AC disease. Moreover, it should also be taken into account that, as demonstrated here, the loss of KIR2DL2 in patients homozygous for KIR2DL3 clearly reduces this protective effect.
Conversely, an increase in the telomeric activating KIR (KIR2DS1, KIR2DS5 and KIR2D3DS1) frequency was observed in AC patients, but this increase only reached statistical significance when the KIR2DS5 genes in total or non-viral AC groups were compared with the controls. Data suggest that in the absence of concomitant viral infection, KIR2DS5 may act as a susceptibility factor for AC disease.
Until now, the ligand of KIR2DS5 receptor has remained unknown, although recently it has been suggested that its association with HLA-C1 may help protect against ankylosing spondylitis or acute kidney graft rejection [56]. However, there is no clear evidence of any interactions between KIR2DS5 and HLA class I molecules. Despite having no known ligand, there are numerous publications that describe the presence and implications of the KIR2DS5 gene in various pathologies [57, 58] and others that suggest more strongly its protective effect in some clinical conditions [59, 60].
This putative effect of KIR2DS5 could be explained by considering that profibrotic cell types involved in the production of extracellular matrix proteins and collagens (myofibroblasts, bone marrow-derived progenitor cells or hepatocytes), as well as Kupffer cells, may express specific ligands for a receptor encoded by this KIR gene. This would have a pro-fibrotic effect through the production of different interferons and cytokines required for HSC activation. From this point of view, it is important to note that it has also been observed that alcohol consumption amplifies the pro-fibrogenic effects of Kupffer cells through gut permeabilization and increasing hepatic endotoxin levels [61, 62], while attenuating the NK-cell anti-fibrotic effect, changes that would accelerate liver fibrosis in patients suffering from viral infections [63]. Taken together, these observations suggest the possibility that a KIR2DS5 ligand is expressed in hepatocytes damaged by alcohol consumption, particularly in advanced stages of AC in patients older than 54 years.
As in the case of the gene pair KIR2DL2/S2, the effect of KIR2DS5 was mainly seen in non-viral alcoholic cirrhotic patients older than 54 years, but not in those aged 54 and under. Perhaps these differences are due to the involvement of different NK cell subtypes, depending on the age, because a decrease in CD56bright NK cells and an expansion of the CD56dim NK subset have been associated with healthy elderly individuals. Additionally, CD57 expression is also higher in elderly populations, which supports the idea that a remodelling process of NK cell subsets may occur during aging, associated with a gradual decrease in the more immature CD56bright NK cells and an increase in highly differentiated CD56dim CD57+ NK cells [64, 65], especially in light of the fact that age itself does not necessarily increase the degree of fibrosis.
In view of the results obtained here, a dual function of NK cells in advanced stages of liver fibrosis in genetically susceptible patients is conceivable. Nevertheless, it should be noted that this retrospective study has certain limitations, mainly related to the fact that patients were asked to declare their alcohol consumption, and their statements did not allow us to extract data on the type of alcohol or the real amount and intensity of their consumption. Thus, although there were no limitations concerning the number of analysed patients for the above-mentioned statistical treatments to be carried out, new prospective studies are needed to determine the impact of these variables and to ratify our results, especially using a larger cohort of viral alcoholic cirrhotic patients. Moreover, these results can only be extrapolated to populations of Caucasian men with AC as the main indication for LT, and with similar demographic characteristics in terms of age, sex and body mass index.
In conclusion, the above data taken together suggest that KIR2DL2 and KIR2DS5 genes behave in opposite ways, KIR2DL2 appearing to act as a protector against the cirrhotic process, whereas KIR2DS5 seems to promote the disease, at least in patients older than 54 years without viral infections. For all these reasons and in view of our results, it would be valuable to know the presence of the KIR2DS5 gene in men with high alcohol consumption to design new strategies to prevent, detect and reduce this liver pathology in these patients and avoid liver transplantation. It would therefore be very interesting to continue this study, with particular attention paid to the expression and function of KIR genes, analysing the interaction with their known (HLA) class I molecule ligands, and also to identify KIR2DS5 receptor ligands in cells that might be involved in the development of AC.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Mann RE, Smart RG, Govoni R. The epidemiology of alcoholic liver disease. Alcohol Res Health. 2003;27:209–19. [PMC free article] [PubMed] [Google Scholar]
- 2.Parker R, Kim SJ, Gao B. Alcohol, adipose tissue and liver disease: mechanistic links and clinical considerations. Nat Rev Gastroenterol Hepatol. 2017;15:50–9. doi: 10.1038/nrgastro.2017.116. [DOI] [PubMed] [Google Scholar]
- 3.Sao R, Aronow WS. Association of non-alcoholic fatty liver disease with cardiovascular disease and subclinical atherosclerosis. Arch Med Sci. 2018;14:1233–44. doi: 10.5114/aoms.2017.68821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao L, Meng F, Cheng J, Li H, Han J, Zhang W. Prediction of oesophageal varices in patients with primary biliary cirrhosis by non-invasive markers. Arch Med Sci. 2017;13:370–6. doi: 10.5114/aoms.2017.65450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo HC, Kuo DP, Chen YL. Impact of beverage consumption, age, and site dependency on dual energy X-ray absorptiometry (DEXA) measurements in perimenopausal women: a prospective study. Arch Med Sci. 2017;13:1178–87. doi: 10.5114/aoms.2017.66033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciećko-Michalska I, Szczepanek M, Wierzbicka-Tutka I, Zahradnik-Bilska J, Mach T. Non-invasive diagnosis of steatosis, inflammatory changes and liver fibrosis in patients with non-alcoholic fatty liver diseases. Pilot study. Arch Med Sci Atheroscler Dis. 2018;3:179–83. doi: 10.5114/amsad.2018.81184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adam R, Karam V, Delvart V, et al. Evolution of indications and results of liver transplantation in Europe. A report from the European Liver Transplant Registry (ELTR) J Hepatol. 2012;57:675–88. doi: 10.1016/j.jhep.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Singal AK, Anand BS. Recent trends in the epidemiology of alcoholic liver disease. Clin Liver Dis. 2013;2:53–6. doi: 10.1002/cld.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legaz I, Navarro-Noguera E, Bolarín JM, et al. Epidemiology, evolution, and long-term survival of alcoholic cirrhosis patients submitted to liver transplantation in southeastern Spain. Alcohol Clin Exp Res. 2016;40:794–805. doi: 10.1111/acer.13013. [DOI] [PubMed] [Google Scholar]
- 10.Burra P, Senzolo M, Adam R, Delvart V, Karam V, Germani G, Neuberger J, ELITA, ELTR Liver Transplant Centers Liver transplantation for alcoholic liver disease in Europe: a study from the ELTR (European Liver Transplant Registry) Am J Transplant. 2010;10:138–48. doi: 10.1111/j.1600-6143.2009.02869.x. [DOI] [PubMed] [Google Scholar]
- 11.Legaz I, Navarro Noguera E, Bolarín JM, et al. Patient sex in the setting of liver transplant in alcoholic liver disease. Exp Clin Transplant. 2019;17:355–62. doi: 10.6002/ect.2017.0302. [DOI] [PubMed] [Google Scholar]
- 12.Suvak B, Dulger AC, Suvak O, Yesilyurt AÖ, Gultepe B, Guducuoglu H. The prevalence and impact of brucellosis in patients with hepatitis delta virus infection: inside the Brucella outbreak with cirrhosis. Arch Med Sci. 2017;13:377–82. doi: 10.5114/aoms.2017.65317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purohit V, Brenner DA. Mechanisms of alcohol-induced hepatic fibrosis: a summary of the Ron Thurman Symposium. Hepatology. 2006;43:872–8. doi: 10.1002/hep.21107. [DOI] [PubMed] [Google Scholar]
- 14.Siegmund SV, Dooley S, Brenner DA. Molecular mechanisms of alcohol-induced hepatic fibrosis. Dig Dis. 2005;23:264–74. doi: 10.1159/000090174. [DOI] [PubMed] [Google Scholar]
- 15.Jelski W, Laniewska-Dunaj M, Orywal K, Kochanowicz J, Rutkowski R, Szmitkowski M. The diagnostic value of alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase (ALDH) measurement in the sera of patients with brain tumor. Arch Med Sci. 2017;13:346–52. doi: 10.5114/aoms.2017.65366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest. 2007;117:539–48. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–18. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao B, Jeong WI, Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47:729–36. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 19.Gao B, Radaeva S, Park O. Liver natural killer and natural killer T cells: immunobiology and emerging roles in liver diseases. J Leukoc Biol. 2009;86:513–28. doi: 10.1189/jlb.0309135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–63. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 21.Miller AM, Horiguchi N, Jeong WI, Radaeva S, Gao B. Molecular mechanisms of alcoholic liver disease: innate immunity and cytokines. Alcohol Clin Exp Res. 2011;35:787–93. doi: 10.1111/j.1530-0277.2010.01399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byun JS, Jeong WI. Involvement of hepatic innate immunity in alcoholic liver disease. Immune Netw. 2010;10:181–7. doi: 10.4110/in.2010.10.6.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao B, Radaeva S, Jeong WI. Activation of natural killer cells inhibits liver fibrosis: a novel strategy to treat liver fibrosis. Expert Rev Gastroenterol Hepatol. 2007;1:173–80. doi: 10.1586/17474124.1.1.173. [DOI] [PubMed] [Google Scholar]
- 24.Pinzani M, Macias-Barragan J. Update on the pathophysiology of liver fibrosis. Expert Rev Gastroenterol Hepatol. 2010;4:459–72. doi: 10.1586/egh.10.47. [DOI] [PubMed] [Google Scholar]
- 25.Marra F, Aleffi S, Galastri S, Provenzano A. Mononuclear cells in liver fibrosis. Semin Immunopathol. 2009;31:345–58. doi: 10.1007/s00281-009-0169-0. [DOI] [PubMed] [Google Scholar]
- 26.Jeong WI, Park O, Radaeva S, Gao B. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology. 2006;44:1441–51. doi: 10.1002/hep.21419. [DOI] [PubMed] [Google Scholar]
- 27.Legaz I, López-Álvarez MR, Campillo JA, et al. KIR gene mismatching and KIR/C ligands in liver transplantation: consequences for short-term liver allograft injury. Transplantation. 2013;95:1037–44. doi: 10.1097/TP.0b013e318286486c. [DOI] [PubMed] [Google Scholar]
- 28.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–7. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 29.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 30.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 31.Dupont B, Selvakumar A, Steffens U. The killer cell inhibitory receptor genomic region on human chromosome 19q13.4. Tissue Antigens. 1997;49:557–63. doi: 10.1111/j.1399-0039.1997.tb02802.x. [DOI] [PubMed] [Google Scholar]
- 32.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–60. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 33.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 34.Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 36.Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heidelbaugh JJ, Bruderly M. Cirrhosis and chronic liver failure: part I. Diagnosis and evaluation. Am Fam Physician. 2006;74:756–62. [PubMed] [Google Scholar]
- 38.Vidal-Castiñeira JR, López-Vázquez A, Martínez-Borra J, et al. Diversity of killer cell immunoglobulin-like receptor (KIR) genotypes and KIR2DL2/3 variants in HCV treatment outcome. PLoS One. 2014;9:e99426. doi: 10.1371/journal.pone.0099426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh KK, Qin M, Brummel SS, et al. Killer cell immunoglobulin-like receptor alleles alter HIV disease in children. PLoS One. 2016;11:e0151364. doi: 10.1371/journal.pone.0151364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozturk OG, Sahın G, Karacor EDZ, Kucukgoz U. Evaluation of KIR genes in recurrent miscarriage. J Assist Reprod Genet. 2012;29:933–8. doi: 10.1007/s10815-012-9811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–22. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 42.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: www.allelefrequencies.net. Tissue Antigens. 2003;61:403–7. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 43.McQueen KL, Dorighi KM, Guethlein LA, Wong R, Sanjanwala B, Parham P. Donor-recipient combinations of group A and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum Immunol. 2007;68:309–23. doi: 10.1016/j.humimm.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyo CW, Guethlein LA, Vu Q, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116:2411–9. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong WI, Gao B. Innate immunity and alcoholic liver fibrosis. J Gastroenterol Hepatol. 2008;23:S112–8. doi: 10.1111/j.1440-1746.2007.05274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Legaz I, Navarro-Noguera E, Bolarín JM, et al. Epidemiology, evolution, and long-term survival of alcoholic cirrhosis patients submitted to liver transplantation in southeastern Spain. Alcohol Clin Exp Res. 2016;40:794–805. doi: 10.1111/acer.13013. [DOI] [PubMed] [Google Scholar]
- 48.Ray S, Khanra D, Sonthalia N, et al. Clinico-biochemical correlation to histological findings in alcoholic liver disease: a single centre study from eastern India. J Clin Diagn Res. 2014;8:MC01–5. doi: 10.7860/JCDR/2014/8763.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfitzmann R, Schwenzer J, Rayes N, Seehofer D, Neuhaus R, Nüssler NC. Long-term survival and predictors of relapse after orthotopic liver transplantation for alcoholic liver disease. Liver Transplant. 2007;13:197–205. doi: 10.1002/lt.20934. [DOI] [PubMed] [Google Scholar]
- 50.Sajja KC, Mohan DP, Rockey DC. Age and ethnicity in cirrhosis. J Investig Med. 2014;62:920–6. doi: 10.1097/JIM.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melhem A, Muhanna N, Bishara A, et al. Anti-fibrotic activity of NK cells in experimental liver injury through killing of activated HSC. J Hepatol. 2006;45:60–71. doi: 10.1016/j.jhep.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 52.Gao B, Radaeva S. Natural killer and natural killer T cells in liver fibrosis. Biochim Biophys Acta. 2013;1832:1061–9. doi: 10.1016/j.bbadis.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radaeva S, Sun R, Jaruga B, Nguyen VT, Tian Z, Gao B. Natural killer cells ameliorate liver fibrosis by killing activated stellate cells in NKG2D-dependent and tumor necrosis factor-related apoptosis-inducing ligand-dependent manners. Gastroenterology. 2006;130:435–52. doi: 10.1053/j.gastro.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 54.Moretta A, Bottino C, Vitale M, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 55.Motavaf M, Safari S, Alavian SM. Therapeutic potential of RNA interference: a new molecular approach to antiviral treatment for hepatitis C. J Viral Hepat. 2012;19:757–65. doi: 10.1111/jvh.12006. [DOI] [PubMed] [Google Scholar]
- 56.Nowak I, Majorczyk E, Wiśniewski A, et al. Does the KIR2DS5 gene protect from some human diseases? PLoS One. 2010;5:e12381. doi: 10.1371/journal.pone.0012381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashouri E, Dabbaghmanesh MH, Rowhanirad S, Bakhshayeshkaram M, Ranjbar Omrani G, Ghaderi A. Activating KIR2DS5 receptor is a risk for thyroid cancer. Hum Immunol. 2012;73:1017–22. doi: 10.1016/j.humimm.2012.07.325. [DOI] [PubMed] [Google Scholar]
- 58.Seymour LA, Nourse JP, Crooks P, et al. The presence of KIR2DS5 confers protection against adult immune thrombocytopenia. Tissue Antigens. 2014;83:154–60. doi: 10.1111/tan.12295. [DOI] [PubMed] [Google Scholar]
- 59.Díaz-Peña R, Blanco-Gelaz MA, Njobvu P, López-Vazquez A, Suárez-Alvarez B, López-Larrea C. Influence of HLA-B*5703 and HLA-B*1403 on susceptibility to spondyloarthropathies in the Zambian population. J Rheumatol. 2008;35:2236–40. doi: 10.3899/jrheum.080395. [DOI] [PubMed] [Google Scholar]
- 60.Płoski R, Luszczek W, Kuśnierczyk P, et al. A role for KIR gene variants other than KIR2DS1 in conferring susceptibility to psoriasis. Hum Immunol. 2006;67:521–6. doi: 10.1016/j.humimm.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 61.McClain CJ, Song Z, Barve SS, Hill DB, Deaciuc I. Recent advances in alcoholic liver disease. IV. Dysregulated cytokine metabolism in alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol. 2004;287:G497–502. doi: 10.1152/ajpgi.00171.2004. [DOI] [PubMed] [Google Scholar]
- 62.Tilg H, Diehl AM. Cytokines in alcoholic and nonalcoholic steatohepatitis. N Engl J Med. 2000;343:1467–76. doi: 10.1056/NEJM200011163432007. [DOI] [PubMed] [Google Scholar]
- 63.Safdar K, Schiff ER. Alcohol and hepatitis C. Semin Liver Dis. 2004;24:305–15. doi: 10.1055/s-2004-832942. [DOI] [PubMed] [Google Scholar]
- 64.Gayoso I, Sanchez-Correa B, Campos C, et al. Immunosenescence of human natural killer cells. J Innate Immun. 2011;3:337–43. doi: 10.1159/000328005. [DOI] [PubMed] [Google Scholar]
- 65.Solana R, Campos C, Pera A, Tarazona R. Shaping of NK cell subsets by aging. Curr Opin Immunol. 2014;29:56–61. doi: 10.1016/j.coi.2014.04.002. [DOI] [PubMed] [Google Scholar]