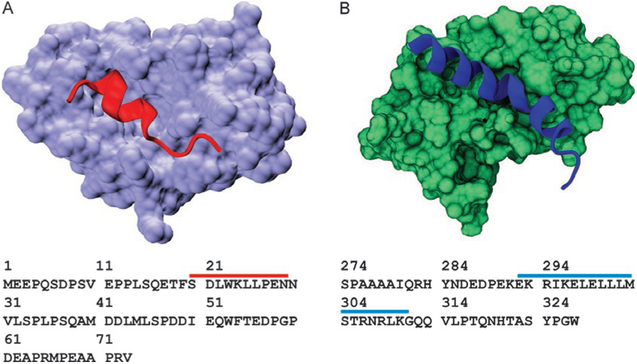
Fig. 3.
Model systems used in this study. (A) Ribbon structure of p53 TAD peptide (red) forming a stable helix when bound to Mdm2 (lavender, PDB ID 1YCR). Sequence of p53 TAD residues 1–73 is shown below the structure. Red bar shows amino acids that fold upon binding to Mdm2. (B) Ribbon structure of c-Myb TAD peptide forming a stable helix when bound to KIX (PDB ID 1SB0). Sequence of c-Myb TAD residues 274–327 is shown below the structure. Blue bar shows amino acids that fold upon binding to KIX.