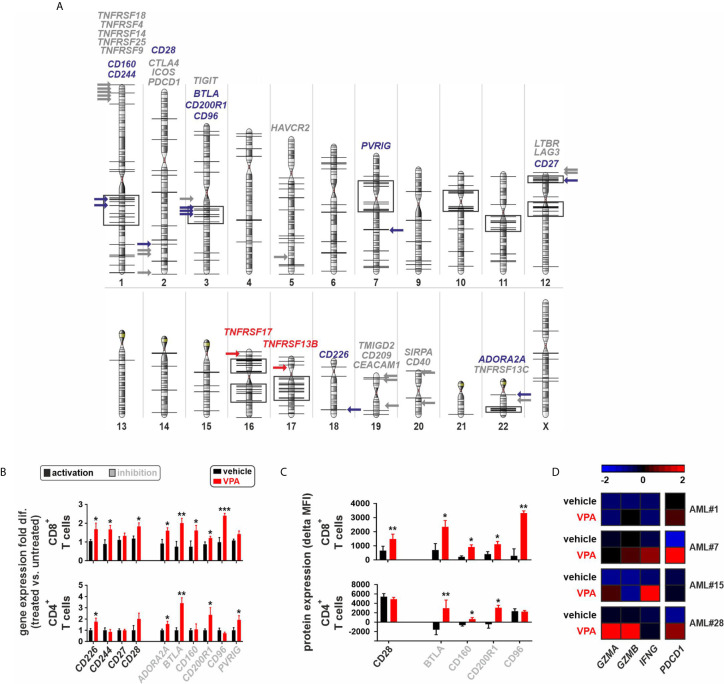
Figure 4.
Epigenetic silencing of immune-checkpoint receptors in BM-infiltrating T cells. (A) Chromosomal position-based gene-mapping (Karyogram) analysis indicating the localization of 30 IC receptors. Black vertical lines on the chromosomal surfaces are indicating the previously identified downregulated genes in CD8+ T cells that were enriched in particular chromosomal regions mainly due to histone deacetylation (12). Black boxes indicate the chromosomal regions containing at least 5 downregulated genes located in a genome distance with less than 5 Mbp from each other, as potential hotspots for the histone remodeling. The 30 newly analyzed IC receptors of AML CD8+ T cells are plotted and highlighted with colored arrows in the karyogram to assess their chromosomal localizations (downregulated ICs: blue arrows; upregulated ICs: red arrows; ICs with no change: gray arrows). (B) Gene expression profile of 10 downregulated IC receptors in CD8+ or CD4+T cells of AML patients upon treatment with VPA. The fold differences were calculated as the ratio of VPA vs. vehicle conditions (n=5 AML patients per each group of CD8+ T cells and n=4 AML patients per each group of CD4+ T cells). (C) Surface protein expression of IC receptors on CD8+ or CD4+ T cells of AML patients. Delta MFI: MFI staining - MFI isotype (n=4 AML patients per group). (D) Heatmap illustrates the expression patterns of three key genes regulating the function of CD8+ T cells as well as PDCD1 gene expression, upon treatment with VPA (n=4 AML patients). Statistics: student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.