Figure 2.
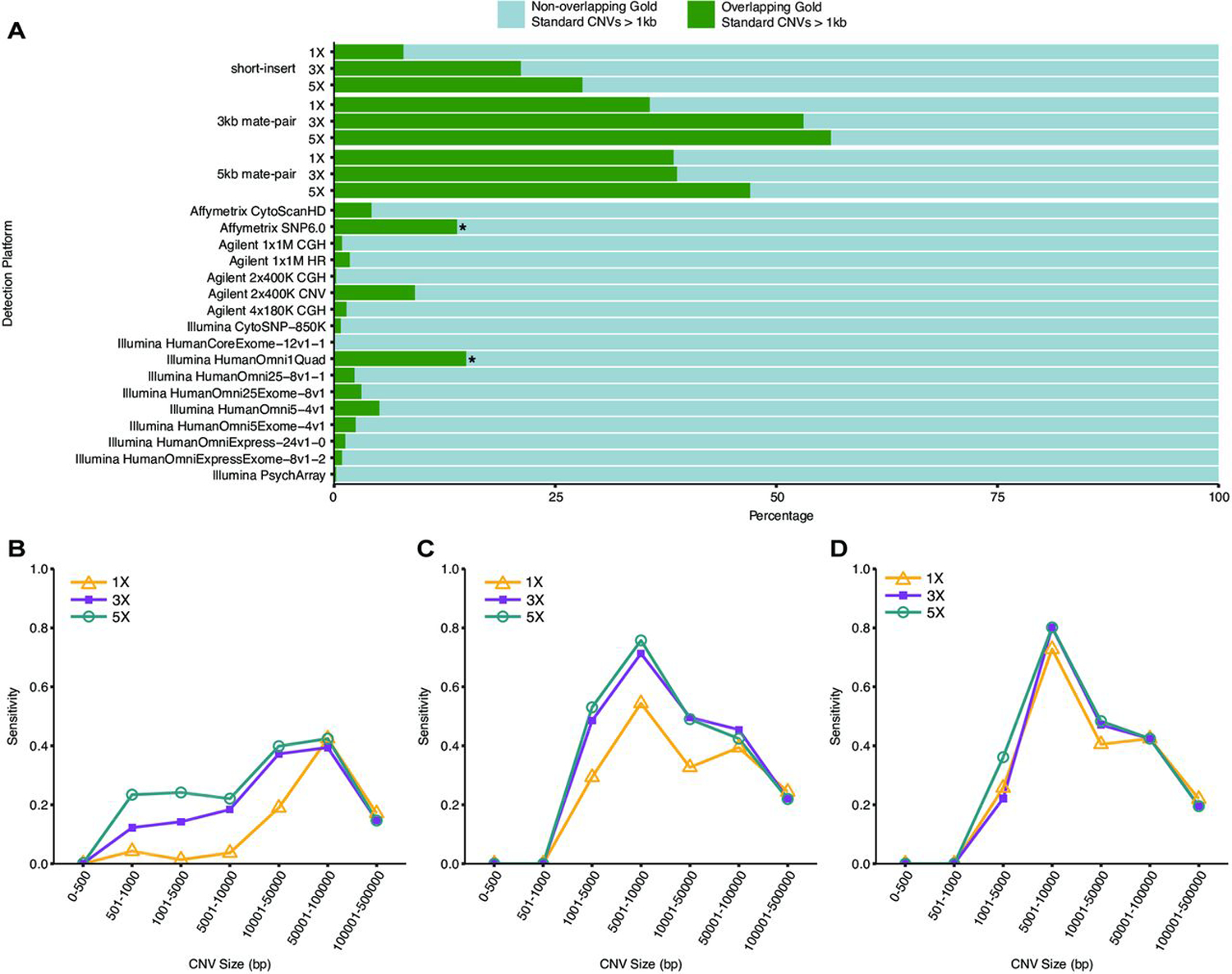
Sensitivity of whole-genome sequencing (WGS) detection of NA12878 GS CNVs (>1 kb). (A) Sensitivity of (>1 kb) GS CNV detection across WGS libraries and array platforms as determined by the ratio of detected autosomal GS CNVs to total number of autosomal GS CNVS. array-based CNV calls were made according to platform-specific algorithms, and WGS CNV calls were made by combining discordant read-pair analysis and read-depth analysis. green: percentage of total autosomal GS CNVs detected (overlapping by >50% reciprocally). light Blue: percentage of total autosomal GS CNVs not detected (non-overlapping by >50% reciprocally). Sensitivity of GS CNV detection in different size ranges from (B) short-insert, (C) 3 kb mate-pair and (D) 5 kb mate-pair libraries at sequencing coverages 1×, 3× and 5×. CNVs were called by combining discordant read-pair analysis and read-depth analysis.
