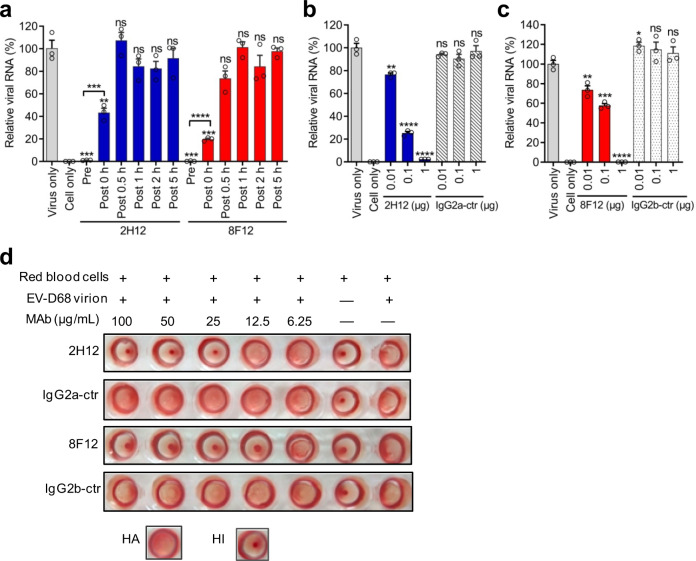
Fig. 3. Anti-EV-D68 MAbs inhibited viral attachment and infection of cells.
a Pre- and post-attachment inhibition assays. 1000 TCID50 of strain 18947 was exposed to the MAbs (2H12 or 8F12) before (pre-attachment, [Pre]) or at different time points after (post-attachment, [Post]) the virus was allowed to adsorb to cooled cells. Total RNA was isolated at 8 h after infection, and EV-D68 RNA was determined by real-time RT-PCR. b, c Inhibition of virus attachment by MAbs 2H12 and 8F12. 1.0 × 107 TCID50 of strains 18953 (b) or 18947 (c) were pre-incubated with various amounts of the indicated antibodies (anti-EV-D68 MAbs 2H12 and 8F12, or isotype control MAbs) for 1 h prior to binding to prechilled RD cells at 4 °C for 1 h. After washing, cells were harvested and total RNA was isolated for real-time RT-PCR analysis of RNA contents of cell-bound virus. For each treatment in panels (a–c), viral RNA levels relative to those for the only virus-infected group are shown. Data are mean ± SEM of triplicate wells. Each symbol represents one well. Statistical significance between virus-only and treated groups was determined by a two-tailed Student’s t-test and was indicated as follows: ns, no significant difference (p ≥ 0.05); *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. In panel (a), p value between the Pre groups and the virus-only group is 0.0001. In panel (b), for the 0.01-μg 2H12 group, p = 0.0042. In panel (c), for the 0.01-μg 8F12 group, p = 0.0084; for the 0.1-μg 8F12 group, p = 0.0005. d Dose-dependent hemagglutination inhibition (HI) by anti-EV-D68 MAbs 2H12 and 8F12. The assay relies on hemagglutination (HA) activity of EV-D68 virion (strain 18947). Symbol (+) indicates presence; (−) indicates absence. In Panels (b–d), ZIKV-specific MAb 1C11 and HCV-specific MAb 1F4 were used as IgG2a and IgG2b isotype controls (ctr), respectively.