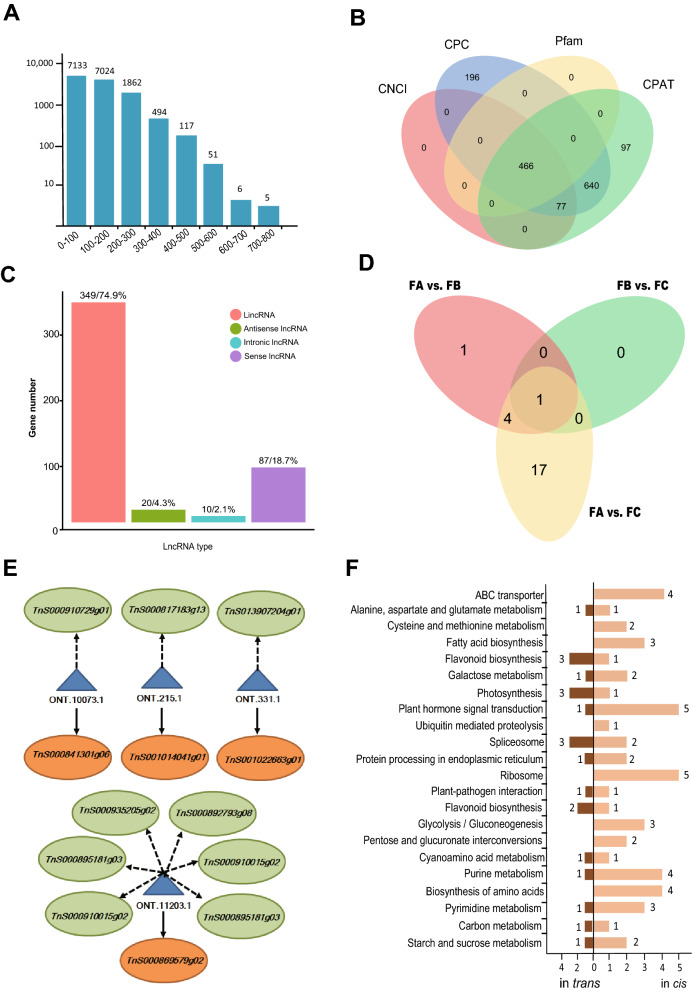
Figure 2.
Identification of open reading frames (ORFs) and lncRNAs based on nine full-length transcriptome. (A) Length distribution of ORFs detected in all full-length transcripts. (B) Venn diagram showing the number of lncRNAs identified using four different approaches: CPC (Coding Potential Calculator), CNCI (Coding-Non-Coding Index), CPAT (Coding Potential Assessment Tool), and Pfam (Protein Family). (C) Functional classification and numbers of four lncRNA types. (D) Venn diagram showing the overlap in the differential expressed lncRNAs among the three different developmental stages of G. luofuense male strobili. (E) Representatives of predicted interaction networks among lncRNAs and their target genes. Solid lines and dotted lines represent the expression regulation by the lncRNAs in cis and in trans, respectively. (F) KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway annotations of cis- and trans-regulated genes regulated by the detected lncRNAs.