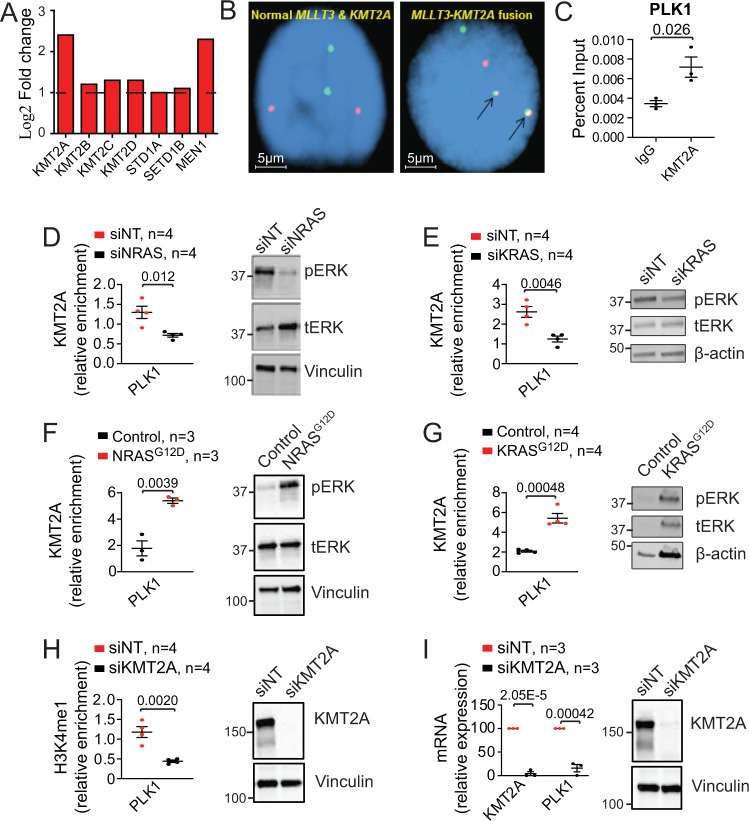
Fig. 6. Mutant RAS regulates PLK1 expression through the lysine methyltransferase KMT2A (MLL1).
A RNA-seq expression levels of enzymes potentially involved in monomethylation in pCMML (vs dCMML and healthy volunteers). B Representative fluorescence in situ hybridization (FISH) in two patients assessing for MLLT3-KMT2A fusions (n = 500 biological replicates). Arrows (right) indicate presence of fusion. C ChIP-PCR in NRAS mutant CMML patient-derived MNCs assessing KMT2A occupancy at the promoter of PLK1 with immunoprecipitation of KMT2A relative to isotype control (IgG). D, E ChIP-PCR assessing KMT2A occupancy at the PLK1 promoter in NRAS mutant (D) and KRAS mutant (E) CMML MNC after transfection with siNT or siNRAS/siKRAS. Western blots are representative validation of RAS knockdown from three experiments. F, G ChIP-PCR assessing KMT2A occupancy at the PLK1 promoter in NRAS wild type (F), and KRAS wild type (G) CMML MNC, after transfection with control vector or NRASG12D/KRASG12D. Western blots are representative validation of RAS expression. H, I ChIP-PCR assessing H3K4me1 at the promoter of PLK1 (H) and qPCR assessing levels of KMT2A and PLK1 (I) in NRAS mutant CMML MNC after transfection with siNT or siKMT2A. Western blots are representative validation of KMT2A knockdown. Western blots in panels D–I are representative of three experiments. Data in panels C–I are presented as mean ± SEM and indicated p-values by two-tailed Student’s t test. The indicated n represents the number of biologic replicates. Source data are provided as a Source Data file.