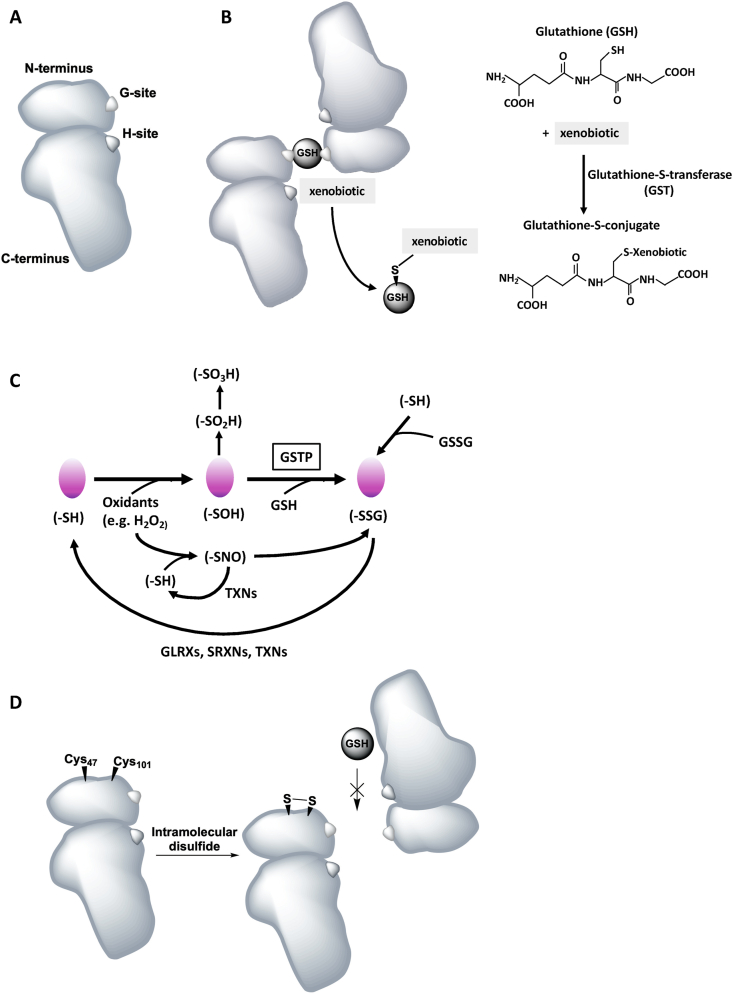
Fig. 2.
Schematic representation of the structure of a Glutathione S-transferase P molecule and the catalytic mechanisms of conjugation of GSTs to xenobiotics and protein S-glutathionylation. A, Schematic of a GSTP monomer with the N- and C-terminal domains including the glutathione binding site, the G-Site, and the xenobiotic binding site, the H-Site. B, Schematic of a GSTP dimer (forming homodimers) binding a GSH molecule on the G-sites in each monomer. Schematic vs molecular representation of the conjugation of GSTS to activated xenobiotics using reduced GSH. C, Catalytic cycle of PSSG and deglutathionylation thought to contribute to the pathogenesis of chronic lung diseases. PSSG can be induced by different biochemical events depending on multiple factors, and can occur spontaneously or is catalyzed by GSTs, notably GSTP. D, GSTP contains cysteines residues regulating its catalytic activity. Intra-subunit disulfide bond formation between Cys47 and Cys101 residues results in steric hindrance for GSH binding. All figures are schematic representations and the actual position of the binding sites and cysteine residues may deviate from the original 3D crystal structure. H2O2: hydrogen peroxide; –SH: protein thiol; –SOH: protein sulfenic acid; –SO2H: protein sulfinic acid; –SO3H: protein sulfonic acid; GSH: glutathione; -SSG: S-glutathionylation; GSSG: glutathione disulfide; –SNO: S-nitrosylation; GLRXs: glutaredoxins; SRNX: sulfiredoxins; TXNs: thioredoxins.