Abstract
Crops are exposed to attacks by various pathogens that cause substantial yield losses and severely threaten food security. To cope with pathogenic infection, crops have elaborated strategies to enhance resistance against pathogens. In addition to the role of protein-coding genes as key regulators in plant immunity, accumulating evidence has demonstrated the importance of non-coding RNAs (ncRNAs) in the plant immune response. Here, we summarize the roles and molecular mechanisms of endogenous ncRNAs, especially microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs), in plant immunity. We discuss the coordination between miRNAs and small interfering RNAs (siRNAs), between lncRNAs and miRNAs or siRNAs, and between circRNAs and miRNAs in the regulation of plant immune responses. We also address the role of cross-kingdom mobile small RNAs in plant–pathogen interactions. These insights improve our understanding of the mechanisms by which ncRNAs regulate plant immunity and can promote the development of better approaches for breeding disease-resistant crops.
Key words: non-coding RNAs, miRNA, lncRNA, circRNA, plant immunity
This review summarizes the roles of miRNAs, siRNAs, lncRNAs, and circRNAs in signal perception, transduction, and downstream responses of plant immunity. Interactions between miRNAs and siRNAs, between lncRNAs and miRNAs or siRNAs, and between circRNAs and miRNAs in the defense against various pathogens are addressed. Finally, the role of cross-kingdom mobile small RNAs in plant–pathogen interactions is discussed.
Introduction
Plants are sessile organisms that are constantly attacked by various pathogens, including bacteria, fungi, and viruses. In the face of these biotic stresses, plants have evolved two layers of immune response: pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Chisholm et al., 2006; Jones and Dangl, 2006; Peng et al., 2018). PAMPs on the cell surface are recognized by plant membrane-located pattern recognition receptors (PRRs) to activate the basal immune response (Jones et al., 2016). ETI is mediated by resistance (R) proteins, commonly intracellular nucleotide-binding/leucine-rich-repeat receptors (NLRs), that specifically recognize their cognate microbial effectors directly or indirectly to induce a robust resistance response (Bialas et al., 2018). Emerging evidence has shown that PTI and ETI share largely overlapping signaling networks and downstream responses (Tsuda et al., 2009; Dodds and Rathjen, 2010; Qi et al., 2011). The concept of three-layer plant immunity has become commonly accepted and involves a recognition layer, a signal-integration layer, and a defense-action layer (Wang et al., 2019). The recognition layer includes PRRs that recognize apoplastic effectors and intracellular receptors that recognize intracellular effectors or host proteins. The signal-integration layer accepts signals from the recognition layer and transmits a set of signals to the defense-action layer. The defense-action layer consists of diverse responses, including the deposition of callose, the production of reactive oxygen species (ROS), and the induction of pathogenesis-related (PR) genes (Wang et al., 2019).
Previous studies have focused on the function of protein-coding genes in biotic stress-response signal transduction. Recent advances in high-throughput gene sequencing technology and large-scale transcriptomic analyses have revealed that a large proportion of the eukaryotic genome is transcribed into RNAs that do not encode proteins. These transcripts are called non-coding RNAs (ncRNAs). Based on their length, ncRNAs are mainly classified into small ncRNAs (sRNAs) (18–30 nt), medium-sized ncRNAs (31–200 nt), and long ncRNAs (lncRNAs) (>200 nt) (Wang et al., 2017a). Plants have two main classes of sRNAs, microRNAs (miRNAs) and small interfering RNAs (siRNAs), which are distinguished by their modes of biogenesis and mechanisms of action. miRNAs are usually 21–24 nt long and are generated from RNAs with imperfectly base-paired hairpin structures. siRNAs are generated from perfectly complementary long double-stranded RNAs (dsRNAs) and may require RNA-dependent RNA polymerases (RDRs) (Borges and Martienssen, 2015). Also, circular RNAs (circRNAs) are a novel type of ncRNA that arises when pre-mRNA is spliced in a reversed order and the 3ʹ and 5ʹ ends are covalently closed (Patop et al., 2019).
Many studies have revealed that ncRNAs play critical roles in plant development, including meristem organization, leaf development, and flowering (Knauer et al., 2013; Csorba et al., 2014; Kidner and Martienssen, 2004; Yang et al., 2019a). Emerging evidence has also shown that ncRNAs are responsive to different environmental stimuli and are implicated in the modulation of gene expression against abiotic and biotic stresses (Hou et al., 2019a; Jabnoune et al., 2013; Li et al., 2018; Kinoshita et al., 2012; Song et al., 2019; Zhao et al., 2018). In this review, we summarize current advances in the regulatory roles of ncRNAs in plant immunity against biotic stress, and we discuss their implications for future studies.
ncRNAs that target regulators of immune perception and transduction
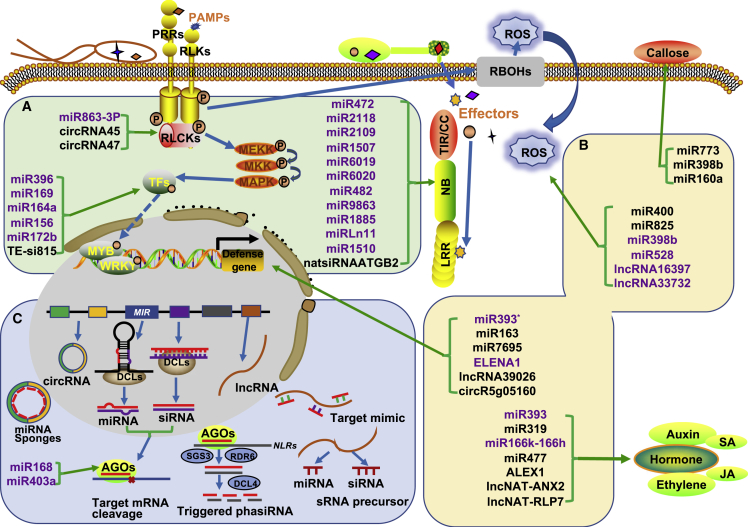
Membrane-located PRRs and cytoplasmic NLRs are two types of immune receptors important to plant immune response (Wang et al., 2019). The membrane-located PRRs comprise mainly receptor-like kinases (RLKs) and receptor-like proteins (RLPs), both of which are usually involved in PTI (Macho and Zipfel, 2014). NLRs are another type of plant immune receptor that is usually involved in ETI (Cui et al., 2015). PPRs and NLRs are regulated at the transcriptional, post-transcriptional, and post-translational levels. The downstream signaling components of these receptors include kinases, transcription factors, and other types of proteins. A number of plant ncRNAs directly target the expression of signaling components to regulate immune response (Figure 1; Table 1).
Figure 1.
ncRNAs involved in plant immunity and their underlying regulation.
(A) ncRNAs that regulate immune signaling components. Some ncRNAs target immune receptor PPRs and NLRs such as FLS2 and RPS5, which mediate pathogen perception. Other ncRNAs regulate RLCKs and TFs such as SpRLK and WRKY45, which are involved in immune signal transduction.
(B) ncRNAs that directly or indirectly modulate various biological processes of PTI or ETI response, including ROS accumulation, callose deposition, defense-related gene expression, and plant hormone regulation, to regulate plant immunity.
(C) Coordinated function among ncRNAs in immunity. miRNAs targeting NLR genes can trigger the production of phasiRNAs. lncRNAs mainly serve as target mimics and miRNA/siRNA precursors. circRNAs may act as decoys for miRNAs to upregulate the expression of mRNAs targeted by miRNAs. Abbreviations: PAMPs, pathogen-associated molecular patterns; PRRs, pattern recognition receptors; RLKs, receptor-like kinases; RLCKs, receptor-like cytosolic kinases; TFs, transcription factors; P, phosphorylation; ROS, reactive oxygen species; DCL, DICER-LIKE; AGO, ARGONAUTE; SGS3, SUPPRESSOR OF GENE SILENCING 3; RDR6, RNA-DEPENDENT RNA POLYMERASE 6; phasiRNA, phased secondary small interfering RNA. ncRNAs marked in purple directly regulate immune signaling components and downstream responses, whereas those marked in black have indirect or unknown mechanisms of regulation. Red lines represent miRNAs and siRNAs.
Table1.
ncRNAs that target plant immune signaling components.
| Function | ncRNAs | Target | Plant species | Reference |
|---|---|---|---|---|
| Trigger phasiRNA production | miR472 | RPS5 | Arabidopsis | Boccara et al. (2014) |
| miR2118/miR2109/miR1507 | Medtr4g023400/Medtr4g014580/Medtr5g071220 | Medicago truncatula | Zhai et al. (2011) | |
| miR6019/miR6020 | N gene | tobacco | Li et al. (2012); Deng et al. (2018b) | |
| miR482 |
FRG3 LRR1/LRR2 EU713768 |
tomato tomato tobacco |
Shivaprasad et al., (2012); Ji et al. (2018); de Vries et al. (2018) | |
| miR9863 | MLA1 | barley | Liu et al. (2014) | |
| Regulate immune receptor | miR1885 | BraTNL1 | Brassica rapa | He et al. (2008); Cui et al. (2020b) |
| miRLn11 | Md-NBS | apple | Ma et al. (2014a) | |
| miR1510 | Glyma.16G135500 | soybean | Cui et al. (2017b) | |
| natsiRNAATGB2 | PPRL | Arabidopsis | Katiyar-Agarwal et al. (2006) | |
| miR172b | TOE1/TOE2 | Arabidopsis | Zou et al. (2018) | |
| lncRNA23468 | miR482b | tomato | Jiang et al. (2019) | |
| lncRNA15492 | miR482a | tomato | Jiang et al. (2020) | |
| Regulate receptor-like kinase | miR863-3P | ARLPK1/ARLPK2 | Arabidopsis | Niu et al. (2016) |
| circRNA45 circRNA47 |
miR477-3P | tomato | Hong et al. (2020) | |
| Transcription factor | miR396 | GRFs | Arabidopsis | Soto-Suarez et al., 2017 |
| miR169 | NF-YAs | rice | Li et al. (2017) | |
| miR164a | NAC60 | rice | Wang et al. (2018a) | |
| miR156 | IPA1 | rice | Liu et al. (2019) | |
| TE-siR815 | ST1 | rice | Zhang et al. (2016b) | |
| lncRNA42705 lncRNA08711 |
mi159 | tomato | Cui et al. (2020a) |
Abbreviations: RPS5, RESISTANT TO P. SYRINGAE 5; LRR1/2, LEUCINE RICH REPEAT 1/2; MLA1, MILDEW RESISTANCE LOCUS A1; TNL1, TIR-NBS-LRR1; NBS, NUCLEOTIDE-BINDING SITE; PPRL, PENTATRICOPEPTIDE REPEAT-LIKE; TOE1/2, TARGET OF EAT1; ARLPK1/2, ATYPICAL RECEPTOR-LIKE PSEUDOKINASE 1/2; GRFs, GROWTH-REGULATING FACTORs; NF-YAs, NUCLEAR FACTOR Y-As; NAC60, NAM/ATAF/CUC 60; IPA1, IDEAL PLANT ARCHITECTURE1; ST1; siR815 TARGET 1.
R proteins are responsible for the recognition of effector proteins secreted by pathogens to initiate stronger ETI (Zhou and Zhang, 2020). Several miRNAs have been shown to guide the cleavage of R genes, serving to tightly control R gene expression to fine-tune the immune response in Arabidopsis, Leguminosae, Solanaceae, and other species. Most of these miRNAs are 22 nt in length and trigger the production of phased siRNAs (phasiRNAs) from their NLR targets (Fei et al., 2013). For example, in Arabidopsis, miR472 (a variant of the miR482 family) targets a typical NLR gene (RPS5) and promotes its phasiRNA biogenesis to repress ETI-based resistance (Boccara et al., 2014). In Medicago truncatula, miR2118a, miR1507, and miR2109 target the NLR genes Medtr4g023400, Medtr5g071220, and Medtr4g014580, respectively, to trigger widespread phasiRNAs (Zhai et al., 2011). In tobacco, miR6019 and miR6020 guide cleavage of transcripts of the TIR-NB-LRR immune receptor gene N, which confers resistance against tobacco mosaic virus (Li et al., 2012; Deng et al., 2018b). In barley, some members of the miR9863 family target a subset of NLR Mla alleles to confer race-specific disease resistance against the powdery mildew fungus (Liu et al., 2014). Discoveries in different plant species indicate that several miRNAs target multiple NLR motifs, such as TIR1, kinase-2, and P-loop motifs (Fei et al., 2013). For example, the miR482/2118 superfamily triggers the production of phasiRNAs by targeting the conserved P-loop-encoding sequence in NLR genes (Shivaprasad et al., 2012; de Vries et al., 2019). These findings suggest that there is a high level of redundancy in the miRNA-mediated suppression of NLR genes.
Some miRNAs silence NLR genes by mRNA cleavage rather than by triggering phasiRNA production to further suppress NLR genes. For example, in soybean, miR1510 targets the NLR gene 16G135500 for mRNA cleavage and negatively regulates plant resistance against Phytophthora sojae (Cui et al., 2017b). In apple, Md-miRLn11 directly targets the NLR gene Md-NBS for mRNA cleavage and compromises plant resistance against apple leaf spot disease (Ma et al., 2014a). These facts suggest that diverse plant species may use a broadly conserved immunity mechanism in which miRNAs directly target NLR genes to regulate the immune response.
Some miRNAs and siRNAs directly target regulators for signal transduction to modulate immunity. In Arabidopsis, miR172b enhances the transcription of the immune receptor gene FLS2 by targeting TOE1 and TOE2, both of which bind directly to the FLS2 promoter to inhibit its activity (Zou et al., 2018). miR863-3P participates in mRNA cleavage and degradation of two atypical receptor-like pseudokinase genes (ARLPK1 and ARLPK2) that are negative regulators of plant immunity during the early stages of Pseudomonas syringae pv. tomato (Pst) infection in Arabidopsis (Niu et al., 2016). The natural antisense transcript-associated siRNA natsiRNAATGB2 silences a gene encoding a pentatricopeptide repeat protein that functions as a negative regulator of RPS2-mediated ETI during infection by Pst, which carries the avrRpt2 effector gene (Katiyar-Agarwal et al., 2006). In tomato, circRNA45 and circRNA47 may act as positive regulators of immunity against P. infestans by regulating the expression of miR477-3P and SpRLK (Hong et al., 2020). These ncRNAs are elaborate regulators of immune signal transduction to modulate plant immunity.
Many miRNAs and siRNAs target transcription factors to regulate the transcription of defense-related genes. In Arabidopsis, miR396 expression decreases during Plectosphaerella cucumerina infection, allowing its GROWTH-REGULATING FACTOR (GRF) transcription factor targets to trigger the reprogramming of gene expression in the host (Soto-Suarez et al., 2017). In rice, miR169 targets NF-YA transcription factors to negatively regulate plant immunity against Magnaporthe oryzae (Li et al., 2017). miR164a expression is suppressed by M. oryzae infection, derepressing its targeted transcription factor OsNAC60, which subsequently increases ROS production, callose accumulation, and the expression of defense-related genes to enhance rice immunity (Wang et al., 2018). miR156 negatively regulates rice resistance against bacterial blight by targeting the squamosa promoter-binding protein-like transcription factors IPA1 and SPL7 (Liu et al., 2019). The transposable element-encoded siRNA TE-siR815 is generated from the intron of WRKY45-1 and represses ST1, which encodes a leucine-rich repeat receptor kinase-type protein, to attenuate WRKY45-mediated rice resistance against Xanthomonas oryzae pv. oryzae (Xoo) (Zhang et al., 2016b). These ncRNAs mediate transcriptional reprogramming upon pathogen infection by directly targeting transcription factors to regulate diverse immune responses.
ncRNAs that regulate the production of defense markers, hormone biosynthesis and signaling, or RNA-mediated silencing
Plant immune response is a complex biological process. Immune signaling typically includes the production of ROS, activation of the MAPK cascade, reprogramming of gene expression (for example, PR genes), accumulation of secondary metabolites, and regulation of plant hormone biosynthesis and signaling. A number of ncRNAs play critical roles in PTI or ETI responses by regulating various biological processes (Figure 1 and Table 2).
Table 2.
ncRNAs that affect other aspects of the immune response.
| Function | ncRNAs | Target | Plant species | References |
|---|---|---|---|---|
| ROS accumulation | miR400 | PPR1/PPR2 | Arabidopsis | Park et al. (2014) |
| miR825 | At5g38850/At3g04220 | Arabidopsis | Nie et al. (2019) | |
| miR398b | CSD1/CSD2/SODX | Arabidopsis/rice | Li et al. (2014); Li et al. (2019) | |
| miR528 | AO | rice | Wu et al. (2017); Yao et al. (2019) | |
| lncRNA16397 | SIGRX21/SIGRX22 | tomato | Cui et al. (2017a) | |
| lncRNA33732 | RBOH | tomato | Cui et al. (2019) | |
| PR gene expression | miR393∗ | MEMB12 | Arabidopsis | Zhang et al. (2011) |
| miR163 | PXMT1/FAMT | Arabidopsis | Chow and Ng (2017) | |
| miR7695 | Nramp6 | rice | Campo et al. (2013); Sanchez-Sanuy et al. (2019) | |
| ELENA1 | PR1 | Arabidopsis | Seo et al. (2017); Seo et al. (2019) | |
| lncRNA39026 | miR168a | tomato | Hou et al. (2020) | |
| circR5g05160 | rice | Fan et al. (2020) | ||
| Callose deposition | miR773 | MET2 | Arabidopsis | Salvador-Guirao et al. (2018a) |
| miR160a | ARF16 | rice | Li et al. (2010) | |
| Hormone | miR393 | TIR1/AFB2/AFB3 | Arabidopsis | Navarro et al. (2006) |
| miR319 | TCP21 | rice | Zhang et al. (2016a); Zhang et al. (2018b) | |
| miR166k-166h | EIN2 | rice | Salvador-Guirao et al. (2018b) | |
| miR477 | CPB60A | cotton | Hu et al. (2020) | |
| ALEX1 | rice | Yu et al. (2020) | ||
|
GhlncNAT-ANX2 GhlncNAT-RLP7 |
LOX1/LOX2 | cotton | Zhang et al. (2018a) | |
| miRNA biosynthesis pathway | miR863-3P | SERRATE | Arabidopsis | Niu et al. (2016) |
| miR168 | AGO1 | rice | Wu et al. (2015) | |
| miR444 | MADS23/27a/57 | rice | Wang et al. (2016a) | |
| miR403a | AGO2 | tobacco | Diao et al. (2019) |
Abbreviations: PPR1/2, PENTATRICOPEPTIDE REPEAT 1/2; CSD1/2, COPPER/ZINC SUPEROXIDE DISMUTASE 1/2; SODX, SUPEROXIDE DISMUTASE; AO, L-ASCORBATE OXIDASE; GRX21/22, GLUTAREDOXIN 21/22; RBOH, RESPIRATORY BURST OXIDASE HOMOLOG; MEMB12, MEMBRIN 12; FAMT, FARNESOIC ACID METHYLTRANSFERASE; Nramp6, natural resistance-associated macrophage protein 6; PR1, PATHOGENESIS-RELATED 1; MET2, METHYLTRANSFERASE 2; ARF16, AUXIN RESPONSE FACTOR 16; TIR1, TRANSPORT INHIBITOR RESPONSE 1; AFB2/3, AUXIN SIGNALING F-BOX PROTEINS 2/3; TCP21, TEOSINTE BRANCHED 1, CYCLOIDEA, PROLIFERATING CELL NUCLEAR ANTIGEN BINDING FACTOR 21; EIN2, ETHYLENE INSENSITIVE 2; CPB60A, CALMODULIN BINDING PROTEIN 60A; LOX1/2, LIPOXYGENASE 1/2; AGO1/2, ARGONAUTE 1/2; ELENA1, ELF18-INDUCED LONG NONCODING RNA; ALEX1, AN LEAF EXPRESSED AND XOO-INDUCED LNCRNA1; ANX2, ANXUR2; RLP7, RECEPTOR-LIKE PROTEIN 7.
Many ncRNAs are involved in the regulation of ROS production. In Arabidopsis, for example, miR400 guides the cleavage of two pentatricopeptide repeat (PPR) genes, resulting in greater ROS accumulation and impaired disease resistance against Pst and Botrytis cinerea (Park et al., 2014). Compared with wild-type plants, Arabidopsis transgenic plants with attenuated miR825 and miR825∗ levels show higher ROS production and therefore exhibit enhanced resistance against B. cinerea (Nie et al., 2019). In rice, miR398b overexpression reduces the transcript levels of genes encoding superoxide dismutases (CSD1, CSD2, SODX, and CCSD), leading to elevated ROS production and enhanced plant resistance against M. oryzae (Li et al., 2014, 2019). miR528 negatively regulates viral resistance in rice by cleaving L-ascorbate oxidase (AO) mRNAs, thereby reducing ROS accumulation (Wu et al., 2017; Yao et al., 2019). In addition, the tomato lncRNA16397 induces GRX expression to reduce ROS accumulation and cell membrane damage, leading to enhanced resistance against P. infestans (Cui et al., 2017a). In tomato, lncRNA33732 is activated by WRKY1 and induces RBOH expression to increase ROS accumulation during early defense against P. infestans attack (Cui et al., 2019). These examples suggest that ncRNAs regulate ROS accumulation by directly targeting the cleavage of genes that encode synthetase or scavenger enzymes of ROS to modulate plant immunity.
Induction of PR gene expression is observed in many disease-resistant plants. Arabidopsis miR393∗, the complementary strand of miR393 within the miRNA/miRNA∗ duplex, contributes to immunity against Pst (avrRpt2) by targeting the protein trafficking gene Membrin 12 (MEMB12) to promote the secretion of PR1 proteins (Zhang et al., 2011). In the basal defense of Arabidopsis against Pst, miR163 acts as a negative regulator by targeting PXMT1 and FAMT to inhibit the expression of defense genes, including PR1 and NPR1 (Chow and Ng, 2017). Also, an elf18-responsive lncRNA (ELENA1) induces PR1 expression by directly interacting with mediator subunit 19a (MED19a) to enrich MED19a on the PR1 promoter in Arabidopsis (Seo et al., 2017). Furthermore, ELENA1 dissociates the FIB2/MED19a complex and releases FIB2 from the PR1 promoter to enhance PR1 expression, thereby increasing plant resistance against Pst (Seo et al., 2019). In rice, osa-miR7695 targets an iron transporter gene that encodes natural resistance-associated macrophage protein 6 (OsNramp6) and enhances the expression levels of defense-related genes, including PR genes and diterpenoid biosynthetic genes (Campo et al., 2013; Sanchez-Sanuy et al., 2019). In tomato, lncRNA39026 induces the expression of PR genes such as PR1 and PR2 to increase plant resistance against P. infestans (Hou et al., 2020). In addition, the overexpression of a circRNA (circR5g05160) in rice enhances plant resistance against M. oryzae by upregulating defense-related genes, including PR10b, PBZ1, HSP90, and SGT1 (Fan et al., 2020). Thus, many ncRNAs regulate the expression of PR genes indirectly. However, the underlying mechanism is largely unknown.
Callose deposition is a typical PTI response against pathogen infection. In Arabidopsis, the flg22 PAMP can decrease miR773 accumulation and therefore derepress its target gene methyltransferase 2 (MET2), resulting in increased callose deposition and enhanced disease resistance against P. cucumerina, Fusarium oxysporum, and Colletotrichum higginsianum (Salvador-Guirao et al., 2018a). Arabidopsis miR398b can reduce callose deposition and compromise disease resistance against Pst (Li et al., 2010). Conversely, miR160a increases PAMP-induced callose deposition and enhances plant immunity (Li et al., 2010). Regulation of callose deposition by the above miRNAs is an indirect result. Hence, future studies will be necessary to clarify how these miRNAs mediate callose deposition.
Some ncRNAs regulate plant immunity by altering the biosynthesis or signaling of plant hormones, including auxin, ethylene, jasmonic acid (JA), and salicylic acid (SA). In Arabidopsis, miRNA393 is induced by flg22 to downregulate the mRNA levels of F-box auxin receptor genes, including TIR1, AFB2, and AFB3, thereby repressing auxin signaling and restricting bacterial growth (Navarro et al., 2006). In rice, miR166k-166h positively regulates rice immunity during M. oryzae and Fusarium fujikuroi infection by controlling the expression of EIN2, which encodes a critical regulator of ethylene signaling (Salvador-Guirao et al., 2018b). Rice miR319 negatively regulates plant immunity by targeting TCP21, which encodes a transcription factor that promotes the expression of LOX2, LOX5, COI1, and COI2 to affect JA biosynthesis and signaling (Zhang et al., 2016a, 2018b). Furthermore, overexpression of the lncRNA ALEX1 in rice upregulates the expression of JA-responsive genes such as JAZ8 and MYC2 and then increases the endogenous levels of JA to enhance resistance against Xoo (Yu et al., 2020). In cotton, miR477 positively regulates plant resistance against Verticillium dahliae by targeting GhCPB60A, which downregulates GhICS1 expression to suppress SA biosynthesis (Hu et al., 2020). Decreased expression of GhlncNAT-ANX2 and GhlncNAT-RLP7 in cotton increases the expression of LOX1 and LOX2, which are involved in the regulation of the JA pathway, enhancing resistance against V. dahliae and B. cinerea (Zhang et al., 2018a). The ncRNA-mediated regulation of plant immunity by fine-tuning multiple plant hormone pathways allows plants to adapt dynamically and flexibly to diverse environments.
The biosynthetic pathways of miRNAs and siRNAs and the components of the RNA-induced silencing complex (RISC) also modulate plant immune response. In Arabidopsis, miR863-3P inhibits the expression of SERRATE, which is essential for the processing of primary miRNA transcripts to miRNAs, thereby suppressing immune response by inhibiting the translation of mRNA (Niu et al., 2016). In rice, stable transgenic expression of Argonaute 18 (AGO18), a key component of the RNA-silencing machinery, elevates the level of AGO1 by sequestering miR168, and therefore confers broad-spectrum virus resistance (Wu et al., 2015). In rice, miR444 upregulates OsRDR1 expression, which is required for the amplification of single-stranded RNAs into dsRNAs, by silencing the MADS-box protein transcriptional repressors of OsRDR1, leading to enhanced virus resistance (Wang et al., 2014, 2016a). In tobacco, miR403a attenuates tobacco mosaic virus resistance by regulating the expression of its target gene AGO2, which enables virus-derived siRNAs to load into the RISC and directs the degradation of the viral RNAs (Diao et al., 2019). Disruption of miRNA biosynthesis and RISC function have a broad impact on diverse aspects of plant immune responses.
Coordinated function among miRNAs, siRNAs, lncRNAs, and circRNAs in plant immunity
Recent studies have demonstrated that a number of ncRNAs alter their expression levels under biotic and abiotic stresses. These ncRNAs, including miRNAs, siRNAs, lncRNAs, and circRNAs, usually interact with each other and coregulate the expression of target genes to modulate plant immunity (Figure 1). The complex interaction among miRNAs, siRNAs, lncRNAs, and circRNAs can coordinate various immune responses in a synergistic or antagonistic manner.
miRNAs and siRNAs
miRNAs and siRNAs are generated by ribonuclease III-like enzyme Dicer or Dicer-like (DCL) proteins and are incorporated into AGO proteins to induce gene silencing by binding to target mRNAs in a sequence-complementary manner (Wu et al., 2009). miRNAs can guide the cleavage of their target mRNAs and trigger the production of secondary siRNAs from their target genes in a phased pattern through the coordinated action of RDR6, Suppressor of Gene Silencing 3, Silencing Defective 5, DCL4, and AGO1 (Peragine et al., 2004; Allen et al., 2005; Hernandez-Pinzon et al., 2007; Song et al., 2012a, 2012b). These secondary siRNAs include mainly trans-acting siRNAs (tasiRNAs) and phasiRNAs, which have a dramatic impact on gene regulation mediated by miRNAs. tasiRNAs act in trans to suppress the expression of genes that are distinct from their original miRNA targets (Vazquez et al., 2004). phasiRNAs are generated from protein-coding genes called PHAS genes and usually promote the cleavage of their target genes in cis. Notably, a set of miRNAs, especially those 22 nt in length that target NLR genes, can trigger the production of phasiRNAs from their target mRNAs. These phasiRNAs further target NLR genes for cleavage to suppress NLR gene function (Fei et al., 2013). These miRNAs are highly conserved in different species, and most of them belong to the miR482/2118 superfamily (Canto-Pastor et al., 2019). This miRNA-NLR-siRNA regulatory mode is an effective way for plants to prevent the autoimmunity and growth inhibition caused by unregulated R gene expression in the absence of pathogens (Deng et al., 2018a). phasiRNAs derived from conserved regions of the NLR genes increase the number of genes targeted by a single miRNA, thus enhancing its silencing effect on NLR genes (Fei et al., 2013). In addition, some miRNAs can target more than one gene to trigger different secondary siRNAs. For instance, in Brassica rapa, the NLR gene BraTNL1 and the trans-acting silencing gene BraTIR1 are both directly targeted by miR1885. phasiRNAs derived from BraTNL1 further target BraTNL1 to regulate plant resistance against TuMV. However, tasiRNAs derived from BraTIR1 target BraCP24 to modulate floral transition (Cui et al., 2020b). Therefore, these 22-nt miRNAs establish regulatory cascades in which mRNAs are targeted by miRNAs and secondary siRNAs.
lncRNAs and miRNAs or siRNAs
Several lines of evidence have shown that some lncRNAs serve as precursors for miRNAs and siRNAs and assist in target gene cleavage. In wheat, three powdery mildew-responsive lncRNAs (TalnRNA5, TapmlnRNA19, and TapmlnRNA8) act as precursors for miRNAs (Xin et al., 2011). Likewise, 16 powdery mildew-responsive lncRNAs act as siRNA precursors, and most of them produce more than one siRNA family. In Brassica napus, 41 lncRNAs that are responsive to Sclerotinia sclerotiorum infection have been identified as precursors for miRNAs, including miR156 and miR169, which play important roles in plant immunity (Joshi et al., 2016). Recently, in mulberry, the novel lncRNA MuLnc1 was found to be cleaved by mul-miR3954 to produce secondary siRNAs, including si161579. It has been demonstrated that si161579 can silence the calmodulin-like protein gene CML27 (MuCML27) in mulberry. MuCML27-overexpressing Arabidopsis plants exhibit enhanced resistance against B. cinerea and Pst. Thus, the miR3954-MuLnc1-siRNAs-mRNAs module provides novel insights into the mulberry defense response (Gai et al., 2018). In addition, it has been reported that lncRNAs containing long-stem structures can be an endogenous source for the generation of DCL1-dependent siRNAs (Ma et al., 2014b). Therefore, to investigate the function of lncRNAs as precursors for miRNAs and siRNAs involved in plant immunity, we must consider the secondary structures of the lncRNAs and the corresponding expression patterns of their cognate miRNAs and siRNAs. It is worth noting that one lncRNA can produce more than one miRNA and/or one siRNA, thus expanding and strengthening the regulation of their target genes.
In addition, several lncRNAs compete with endogenous RNAs (ceRNAs) to sequester miRNAs in a type of target mimicry. In tomato, slylnc0195 and slylnc1077 act as decoys for miR166 and miR399, respectively, to regulate plant resistance against tomato yellow leaf curl virus (TYLCV) (Wang et al., 2015). Similarly, lncRNA39026 in tomato acts as an endogenous target mimic for miR168a to positively modulate defense response to P. infestans infection through the regulation of SlAG O 1 expression (Hou et al., 2020). Also, lncRNA42705 and lncRNA08711 increase the mRNA levels of MYB genes by acting as decoys for miR159, thereby enhancing resistance against P. infestans (Cui et al., 2020a). Overexpression of lncRNA23468, which contains conserved endogenous target mimic sites for miR482b, decreases miR482b expression in tomato, increasing expression of the target NLR gene Solyc02g036270.2 and enhancing resistance against P. infestans (Jiang et al., 2019). The half-life of miRNA is usually very long, making it difficult to rapidly reduce the abundance of miRNA when plants are under pathogen attack. Thus, plants use lncRNAs as endogenous target mimics to repress miRNA activity, which may be faster than inhibiting miRNA biogenesis and enhancing miRNA turnover.
Conversely, an miRNA or siRNA may directly target an lncRNA to attenuate its presence through cleavage. Recently, it has been reported that tomato Sl-lncRNA15492 interacts with Sl-miR482a to regulate host immunity against P. infestans: Sl-lncRNA15492 inhibits the production of mature Sl-miR482a through an Sl-miR482a precursor contained in the antisense sequence of Sl-lncRNA15492. In turn, Sl-miR482a suppresses Sl-lncRNA15492 expression through direct cleavage. It has been noted that Sl-miR482a targets the NLR gene Sl-NBS-LRR1 to negatively regulate plant resistance in tomato. Therefore, Sl-lncRNA15492 and Sl-miR482a mutually inhibit each other to maintain an appropriate Sl-NBS-LRR1 level during regulation of the tomato immune response to P. infestans (Jiang et al., 2020). This interaction between an miRNA and an lncRNA to regulate an NLR gene reveals a novel mechanism for the regulation of plant immunity. In addition, a TYLCV-derived siRNA induces silencing of the tomato lncRNA SlLNR1, which contributes to TYLCV resistance, through the direct cleavage of SlLNR1 (Yang et al., 2019b). The interaction between viral siRNAs and host lncRNAs provides integrative regulation of the arms race between pathogens and hosts.
circRNAs and miRNAs
Although a few cases in animals have shown that circRNAs act as miRNA sponges, the same function for plant circRNAs requires further empirical validation. Bioinformatic analysis has shown that the majority of circRNAs do not possess multiple binding sites for miRNAs and thus may not function as miRNA sponges (Guo et al., 2014). Genome-wide analysis of circRNAs in Arabidopsis and rice has revealed that only a small portion of circRNAs (5.0% in Arabidopsis and 6.6% in rice) are potential target mimics of miRNAs (Ye et al., 2015). As one example, circRNA Os08circ16564 was predicted to be a target mimic of miR172. However, RT–PCR results showed that the miR172 levels in Os08circ16564-overexpressing plants were not significantly different from those in wild-type plants, indicating that Os08circ16564 may not function as a sponge for miR172 (Lu et al., 2015). This result also suggests that serving as miRNA sponges may not be the major mode of circRNA function in plants. However, bioinformatic analysis predicted that 24 circRNAs in tomato and 6 circRNAs in wheat may act as miRNA sponges (Zuo et al., 2016; Wang et al., 2017b). This prediction is largely dependent on linear sequence matching using bioinformatic tools. However, the secondary structures of circRNAs may hide a portion of potential miRNA binding sites, a fact that is often overlooked when predicting potential miRNA binding sites for circRNAs in plants. This may explain why Os08circ16564 overexpression did not affect miR172 levels. In addition, to act as miRNA sponges, each circRNA must have a sufficient number of miRNA binding sites or be present at a sufficiently high level (Ebert and Sharp 2010). Nevertheless, although the mammalian circRNAs ciRS-7 and CDR1 possess high-density miRNA binding sites (Hansen et al., 2013; Memczak et al., 2013), no plant circRNAs contain such a high density of miRNA binding sites. On the other hand, a recent study showed that circRNA45 and circRNA47 levels were induced upon P. infestans infection, and these circRNAs may act as sponges for miR477-3P to regulate its target gene SpRLK in tomato (Hong et al., 2020). Therefore, whether plant circRNAs function to directly inhibit miRNAs requires further investigation. One benefit of circRNAs acting as miRNA sponges may be that one circRNA can target a number of miRNAs simultaneously. When needed, many miRNAs can be quickly released from the cirRNA and miRNA interaction complex to modulate plant immunity rapidly and efficiently.
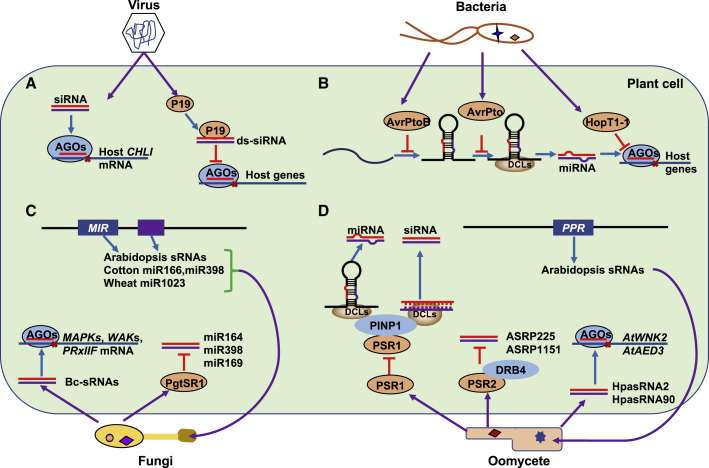
Roles of pathogen effectors and mobile ncRNAs in the interaction between pathogens and host plants.
In addition to host-derived ncRNAs that regulate the biosynthesis and signaling of ncRNAs involved in plant immunity, effectors secreted by distinct pathogens also hijack the RNA-silencing machinery to interfere with plant immunity and achieve successful infection (Figure 2). This type of effector is called an RNA-silencing suppressor, and such suppressors are expressed by pathogenic viruses, bacteria, fungi, and oomycetes (Spanu, 2015; de Vries et al., 2019). For example, the virus P19 protein of tombusviruses prevents post-transcriptional gene silencing by specifically binding to double-stranded siRNAs in tobacco (Silhavy et al., 2002). In bacteria, the HopT1-1 effector of Pst interferes with AGO1 to suppress RNA silencing and cause disease in Arabidopsis (Navarro et al., 2008). In fungi, the PgtSR1 effector proteins from the wheat stem rust pathogen Puccinia graminis f. sp. tritici (Pgt) were the first identified RNA-silencing suppressors from fungi. They compromise wheat immunity by altering the abundance of small RNAs involved in defense processes, including miR164, miR398, and miR169 (Yin et al., 2019). In oomycetes, PSR1 and PSR2 from P. sojae suppress RNA silencing in plants by inhibiting the biogenesis of small RNAs such as miR393 and ASRP255 to promote infection (Qiao et al. 2013, 2015). These results demonstrate that pathogens use a conserved strategy to inhibit plant defense responses by suppressing RNA silencing in the host.
Figure 2.
Effectors and mobile small RNAs in the interactions between pathogens and host plants.
(A) Role of sRNAs in plant–virus interaction. Virus-derived siRNAs directly target a chlorophyll biosynthetic gene in the plant. The virus also encodes proteins, such as P19, to suppress RNA silencing by interfering with DCL function, the binding of virus-derived siRNAs, and the prevention of RISC assembly.
(B) Role of sRNAs in plant–bacteria interaction. Bacterial effectors translocate into host plant cells and inhibit the regulation of plant sRNA. The effector AvrPtoB represses the accumulation of miR393 at the transcriptional level, whereas AvrPto reduces the processing of miR393. HopT1-1 suppresses AGO1 function to inhibit RNA silencing.
(C) Role of sRNAs in plant–fungi interaction. Fungal sRNAs, Bc-sRNAs, translocate into host cells and utilize plant RNA-silencing components to reduce the expression of host immune genes such as MAPKs and WAKs. The fungal effector PgtSR1 regulates the abundance of a set of miRNAs involved in plant immunity. Plants also send several small RNAs into fungi to modulate their growth and virulence.
(D) Role of sRNAs in plant–oomycete interaction. The PSR1 and PSR2 effectors of oomycetes are secreted into plant cells and alter the host RNA-silencing machinery to decrease host immunity. Oomycetes also send small RNAs to silence host genes such as AtWNK2 and AtAED3. Host plants export many small RNAs into oomycetes to change pathogen virulence. Abbreviations: PSR1/2, Phytophthora suppressors of RNA silencing 1/2; DRB4, dsRNA-binding protein 4; PINP1, PSR1-interacting protein 1; DCL, DICER-LIKE; AGO, ARGONAUTE; PPR, pentatricopeptide repeat; AtWNK2, Arabidopsis WITH NO LYSINE (K) KINASE 2; AtAED3, APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT 3; WAKs, cell wall-associated kinases; red lines represent miRNAs and siRNAs.
Notably, regulation by ncRNAs is not restricted to the individual organism in which they are generated. Many mobile miRNAs and siRNAs can translocate between hosts and pathogens, silencing genes in the interacting organism by a mechanism termed cross-kingdom RNAi (Huang et al., 2019) (Figure 2). However, lncRNAs and circRNAs involved in plant immunity have not yet been found to move between pathogens and plants.
Small RNAs derived from pathogens mediate cross-kingdom RNAi in plant hosts and represent a novel class of pathogen effectors that inhibit host immunity for successful infection (Kwon et al., 2020). For example, cucumber mosaic virus Y-satellite RNA-derived siRNAs directly target the chlorophyll biosynthetic gene CHLI in tobacco to cause yellowing symptoms (Smith et al., 2011). In fungi, some small RNAs from Botrytis cinerea, such as Bc-sR3.1, Bc-sR3.2, Bc-sR5, and Bc-sR37, are delivered into host cells and silence Arabidopsis and tomato genes involved in immunity, including MAPKs, cell-wall-associated kinases, and other defense proteins (Weiberg et al., 2013; Wang et al., 2017c). A group of small RNAs from V. dahliae also move into plant cells to silence genes involved in immunity by associating with AGO1 (Wang et al., 2016b). In oomycetes, HpasRNA2 and HpasRNA90, two small RNAs from Hyaloperonospora arabidopsidis, are translocated into Arabidopsis cells and associate with AGO1 to silence the plant defense genes AtWNK2 and AtAED3, respectively (Dunker et al., 2020). This phenomenon has been discovered in several cases of plant–pathogen interactions, indicating that pathogen-induced gene silencing is widespread and complex.
Plant hosts also transport small RNAs into pathogens to suppress the expression of virulence-related genes, thereby contributing to plant defense response (Zhu et al., 2019). Arabidopsis cells secrete exosome-like extracellular vesicles to deliver small RNAs into the fungal pathogen B. cinerea to induce the silencing of fungal genes involved in pathogenicity (Cai et al., 2018). The wheat miRNA1023 inhibits a Fusarium graminearum alpha/beta hydrolase gene that plays an important role in fungal infection (Jiao and Peng, 2018). Similarly, cotton plants export miR166 and miR159 to the V. dahliae hyphae to separately target two V. dahliae genes that encode a Ca2+-dependent cysteine protease (Clp-1) and an isotrichodermin C-15 hydroxylase (HiC-15), leading to enhanced disease resistance (Zhang et al., 2016c). Interestingly, in the interaction between Phytophthora and Arabidopsis, Phytophthora infection induces a set of secondary siRNAs generated from a plant PPR gene cluster, and these siRNAs potentially silence target genes in Phytophthora to confer resistance. As a counter defense mechanism, the PSR2 effector interferes with secondary siRNA production by associating with dsRNA-binding protein 4, which is involved in secondary siRNA biogenesis (Hou et al., 2019b). These results demonstrate that cross-kingdom RNAi is bidirectional during plant–pathogen interactions.
Concluding remarks and perspectives
An increasing number of plant ncRNAs involved in plant immunity have been identified. However, mechanistic details of immunity regulation by these plant ncRNAs are very limited. Accumulating findings highlight important roles for ncRNAs in the regulation of diverse aspects of plant immunity, including pathogen perception, signal transduction, and downstream immune responses. Furthermore, these ncRNAs play multiple roles in plant immunity through the use of different strategies, including modulation of gene expression, interaction with proteins, and interplay with other ncRNAs. An ncRNA can target more than one gene, and a single ncRNA may thus show pleiotropic effects not only on plant immunity but also on other biological processes, including plant development and response to abiotic stresses. For instance, the Brassica miRNA miR1885 regulates both plant growth and immunity by repressing the expression of the NLR gene BraTNL1 and the photosynthesis-related gene BraCP24, suggesting that miR1885 and its target genes are ideal breeding targets for disease resistance and high yield (Cui et al., 2020b). Conversely, one target gene may be regulated by multiple ncRNAs simultaneously, underscoring the diversity and complexity of plant ncRNA modes of action. Taking advantage of this, in the case of growth attenuation caused by R genes, the introduction of regulatory miRNAs may help to achieve a balance between resistance and crop development.
Although the potential models for plants are similar to those for mammals, it remains to be determined whether ncRNA modes of action are conserved between plants and mammals and whether plant ncRNAs exhibit novel mechanisms. Recent studies have shown that mobile ncRNAs translocate between plants and pathogens to mediate cross-kingdom regulation of plant immunity and pathogen virulence (Weiberg et al., 2013; Wang et al., 2016b; Zhang et al., 2016c; Cai et al., 2018; Huang et al., 2019). It is important to characterize the targets of these mobile ncRNAs and their molecular regulation. The diverse subcellular localizations of ncRNAs may be related to their specific roles in plant cells, and determining the subcellular localization of individual ncRNAs is important for understanding their functions. In addition, as ncRNAs may encode peptides, it will be interesting to determine whether these potential peptides play roles in plant immunity and to uncover their underlying mechanisms. We believe that future studies on ncRNA will provide additional insight into plant immunity and offer effective approaches for the improvement of plant disease resistance, thereby ensuring global food security.
Funding
X.C. was supported by funds from the National Natural Science Foundation of China (NSFC) (31825022 and 31772153). L.S. was supported by grants from the Applied Basic Research Programs of Science and Technology Department from Sichuan Province (2021YJ0494). J.W. was supported by grants from the NSFC (32072043 and 31922066).
Author contributions
This work was supervised by J.W. and X.C. The original draft was prepared by L.S., Y.F., and L.C. and reviewed and edited by L.S. and X.C.
Acknowledgments
We thank Dr. Mawsheng Chern from the University of California at Davis, USA for providing critical reading. No conflict of interest declared.
Published: March 20, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Contributor Information
Jing Wang, Email: jingwang406@sicau.edu.cn.
Xuewei Chen, Email: xwchen88@163.com.
References
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bialas A., Zess E.K., De la Concepcion J.C., Franceschetti M., Pennington H.G., Yoshida K., Upson J.L., Chanclud E., Wu C.H., Langner T. Lessons in effector and NLR biology of plant-microbe systems. Mol. Plant Microbe Interact. 2018;31:34–45. doi: 10.1094/MPMI-08-17-0196-FI. [DOI] [PubMed] [Google Scholar]
- Boccara M., Sarazin A., Thiebeauld O., Jay F., Voinnet O., Navarro L., Colot V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014;10:e1003883. doi: 10.1371/journal.ppat.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Qiao L., Wang M., He B., Lin F.M., Palmquist J., Huang S.D., Jin H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360:1126–1129. doi: 10.1126/science.aar4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S., Peris-Peris C., Sire C., Moreno A.B., Donaire L., Zytnicki M., Notredame C., Llave C., San Segundo B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013;199:212–227. doi: 10.1111/nph.12292. [DOI] [PubMed] [Google Scholar]
- Canto-Pastor A., Santos B., Valli A.A., Summers W., Schornack S., Baulcombe D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. U S A. 2019;116:2755–2760. doi: 10.1073/pnas.1814380116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Chow H.T., Ng D.W. Regulation of miR163 and its targets in defense against Pseudomonas syringae in Arabidopsis thaliana. Sci. Rep. 2017;7:46433. doi: 10.1038/srep46433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Questa J.I., Sun Q., Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U S A. 2014;111:16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- Cui J., Jiang N., Meng J., Yang G., Liu W., Zhou X., Ma N., Hou X., Luan Y. LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato-Phytophthora infestans interactions. Plant J. 2019;97:933–946. doi: 10.1111/tpj.14173. [DOI] [PubMed] [Google Scholar]
- Cui J., Luan Y., Jiang N., Bao H., Meng J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017;89:577–589. doi: 10.1111/tpj.13408. [DOI] [PubMed] [Google Scholar]
- Cui X., Yan Q., Gan S., Xue D., Dou D., Guo N., Xing H. Overexpression of gma-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene. 2017;621:32–39. doi: 10.1016/j.gene.2017.04.015. [DOI] [PubMed] [Google Scholar]
- Cui J., Jiang N., Hou X., Wu S., Zhang Q., Meng J., Luan Y. Genome-wide identification of lncRNAs and analysis of ceRNA networks during tomato resistance to Phytophthora infestans. Phytopathology. 2020;110:456–464. doi: 10.1094/PHYTO-04-19-0137-R. [DOI] [PubMed] [Google Scholar]
- Cui C., Wang J.J., Zhao J.H., Fang Y.Y., He X.F., Guo H.S., Duan C.G. A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol. Plant. 2020;13:231–245. doi: 10.1016/j.molp.2019.11.010. [DOI] [PubMed] [Google Scholar]
- de Vries S., de Vries J., Rose L.E. The elaboration of miRNA regulation and gene regulatory networks in plant -microbe interactions. Genes (Basel) 2019;10:310. doi: 10.3390/genes10040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S., Kukuk A., von Dahlen J.K., Schnake A., Kloesges T., Rose L.E. Expression profiling across wild and cultivated tomatoes supports the relevance of early miR482/2118 suppression for Phytophthora resistance. Proc. Biol. Sci. 2018;285:20172560. doi: 10.1098/rspb.2017.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Muhammad S., Cao M., Wu L. Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 2018;16:965–975. doi: 10.1111/pbi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Wang J., Tung J., Liu D., Zhou Y., He S., Du Y., Baker B., Li F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018;14:e1006756. doi: 10.1371/journal.ppat.1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao P., Zhang Q., Sun H., Ma W., Cao A., Yu R., Wang J., Niu Y., Wuriyanghan H. miR403a and SA are involved in NbAGO2 mediated antiviral defenses against TMV infection in Nicotiana benthamiana. Genes (Basel) 2019;10:526. doi: 10.3390/genes10070526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dunker F., Trutzenberg A., Rothenpieler J.S., Kuhn S., Prols R., Schreiber T., Tissier A., Kemen A., Kemen E., Huckelhoven R. Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. eLife. 2020;9:e56096. doi: 10.7554/eLife.56096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Sharp P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Quan W., Li G.B., Hu X.H., Wang Q., Wang H., Li X.P., Luo X., Feng Q., Hu Z.J. circRNAs are involved in the rice-Magnaporthe oryzae interaction. Plant Physiol. 2020;182:272–286. doi: 10.1104/pp.19.00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Y.P., Yuan S.S., Zhao Y.N., Zhao H.N., Zhang H.L., Ji X.L. A novel LncRNA, MuLnc1, associated with environmental stress in mulberry (Morus multicaulis) Front Plant Sci. 2018;9:669. doi: 10.3389/fpls.2018.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- He X.F., Fang Y.Y., Feng L., Guo H.S. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008;582:2445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Hernandez-Pinzon I., Yelina N.E., Schwach F., Studholme D.J., Baulcombe D., Dalmay T. SDE5, the putative homologue of a human mRNA export factor, is required for transgene silencing and accumulation of trans-acting endogenous siRNA. Plant J. 2007;50:140–148. doi: 10.1111/j.1365-313X.2007.03043.x. [DOI] [PubMed] [Google Scholar]
- Hong Y.H., Meng J., Zhang M., Luan Y.S. Identification of tomato circular RNAs responsive to Phytophthora infestans. Gene. 2020;746:144652. doi: 10.1016/j.gene.2020.144652. [DOI] [PubMed] [Google Scholar]
- Hou J., Lu D., Mason A.S., Li B., Xiao M., An S., Fu D. Non-coding RNAs and transposable elements in plant genomes: emergence, regulatory mechanisms and roles in plant development and stress responses. Planta. 2019;250:23–40. doi: 10.1007/s00425-019-03166-7. [DOI] [PubMed] [Google Scholar]
- Hou Y., Zhai Y., Feng L., Karimi H.Z., Rutter B.D., Zeng L., Choi D.S., Zhang B., Gu W., Chen X. A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe. 2019;25:153–165. doi: 10.1016/j.chom.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Cui J., Liu W., Jiang N., Zhou X., Qi H., Meng J., Luan Y. LncRNA39026 enhances tomato resistance to Phytophthora infestans by decoying miR168a and inducing PR gene expression. Phytopathology. 2020;110:873–880. doi: 10.1094/PHYTO-12-19-0445-R. [DOI] [PubMed] [Google Scholar]
- Hu G., Hao M., Wang L., Liu J., Zhang Z., Tang Y., Peng Q., Yang Z., Wu J. The cotton miR477-CBP60A module participates in plant defense against Verticillium dahliae. Mol. Plant Microbe Interact. 2020;33:624–636. doi: 10.1094/MPMI-10-19-0302-R. [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Wang H., Hu P., Hamby R., Jin H. Small RNAs—big players in plant-microbe interactions. Cell Host Microbe. 2019;26:173–182. doi: 10.1016/j.chom.2019.07.021. [DOI] [PubMed] [Google Scholar]
- Jabnoune M., Secco D., Lecampion C., Robaglia C., Shu Q., Poirier Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell. 2013;25:4166–4182. doi: 10.1105/tpc.113.116251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H.M., Zhao M., Gao Y., Cao X.X., Mao H.Y., Zhou Y., Fan W.Y., Borkovich K.A., Ouyang S.Q., Liu P. FRG3, a target of slmiR482e-3p, provides resistance against the fungal pathogen Fusarium oxysporum in tomato. Front. Plant Sci. 2018;9:26. doi: 10.3389/fpls.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Cui J., Hou X., Yang G., Xiao Y., Han L., Meng J., Luan Y. Sl-lncRNA15492 interacts with Sl-miR482a and affects Solanum lycopersicum immunity against Phytophthora infestans. Plant J. 2020;103:1561–1574. doi: 10.1111/tpj.14847. [DOI] [PubMed] [Google Scholar]
- Jiang N., Cui J., Shi Y., Yang G., Zhou X., Hou X., Meng J., Luan Y. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic. Res. 2019;6:28. doi: 10.1038/s41438-018-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J., Peng D. Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J. Plant Interact. 2018;13:514–521. [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Vance R.E., Dangl J.L. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Joshi R.K., Megha S., Basu U., Rahman M.H., Kav N.N. Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS One. 2016;11:e0158784. doi: 10.1371/journal.pone.0158784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A., Jr., Zhu J.K., Staskawicz B.J., Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U S A. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Kinoshita N., Wang H., Kasahara H., Liu J., Macpherson C., Machida Y., Kamiya Y., Hannah M.A., Chua N.H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell. 2012;24:3590–3602. doi: 10.1105/tpc.112.097006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S., Holt A.L., Rubio-Somoza I., Tucker E.J., Hinze A., Pisch M., Javelle M., Timmermans M.C., Tucker M.R., Laux T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell. 2013;24:125–132. doi: 10.1016/j.devcel.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Kwon S., Tisserant C., Tulinski M., Weiberg A., Feldbrugge M. Inside-out: from endosomes to extracellular vesicles in fungal RNA transport. Fungal Biol. Rev. 2020;34:89–99. [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. microRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U S A. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Li Y., Cao X.L., Zhu Y., Yang X.M., Zhang K.N., Xiao Z.Y., Wang H., Zhao J.H., Zhang L.L., Li G.B. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019;222:1507–1522. doi: 10.1111/nph.15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu Y.G., Shi Y., Wu L., Xu Y.J., Huang F., Guo X.Y., Zhang Y., Fan J., Zhao J.Q. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014;164:1077–1092. doi: 10.1104/pp.113.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Zhang J., Wu L., Qi Y., Zhou J.M. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhao S.L., Li J.L., Hu X.H., Wang H., Cao X.L., Xu Y.J., Zhao Z.X., Xiao Z.Y., Yang N. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front Plant Sci. 2017;8:2. doi: 10.3389/fpls.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cheng X., Liu D., Xu W., Wise R., Shen Q.H. The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 2014;10:e1004755. doi: 10.1371/journal.pgen.1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Shi Z., Zhang X., Wang M., Zhang L., Zheng K., Liu J., Hu X., Di C., Qian Q. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants. 2019;5:389–400. doi: 10.1038/s41477-019-0383-2. [DOI] [PubMed] [Google Scholar]
- Lu T., Cui L., Zhou Y., Zhu C., Fan D., Gong H., Zhao Q., Zhou C., Zhao Y., Lu D. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Lu Y., Bai S., Zhang W., Duan X., Meng D., Wang Z., Wang A., Zhou Z., Li T. Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Golden Delicious) Mol. Plant. 2014;7:218–230. doi: 10.1093/mp/sst101. [DOI] [PubMed] [Google Scholar]
- Ma X., Shao C., Jin Y., Wang H., Meng Y. Long non-coding RNAs: a novel endogenous source for the generation of Dicer-like 1-dependent small RNAs in Arabidopsis thaliana. RNA Biol. 2014;11:373–390. doi: 10.4161/rna.28725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A., Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol. Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Navarro L., Jay F., Nomura K., He S.Y., Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie P., Chen C., Yin Q., Jiang C., Guo J., Zhao H., Niu D. Function of miR825 and miR825∗ as negative regulators in Bacillus cereus AR156-elicited systemic resistance to Botrytis cinerea in Arabidopsis thaliana. Int. J. Mol. Sci. 2019;20:5032. doi: 10.3390/ijms20205032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D., Lii Y.E., Chellappan P., Lei L., Peralta K., Jiang C., Guo J., Coaker G., Jin H. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 2016;7:11324. doi: 10.1038/ncomms11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Lee H.J., Kwak K.J., Lee K., Hong S.W., Kang H. MicroRNA400-guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 2014;55:1660–1668. doi: 10.1093/pcp/pcu096. [DOI] [PubMed] [Google Scholar]
- Patop I.L., Wust S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., van Wersch R., Zhang Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 2018;31:403–409. doi: 10.1094/MPMI-06-17-0145-CR. [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Tsuda K., Glazebrook J., Katagiri F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 2011;12:702–708. doi: 10.1111/j.1364-3703.2010.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Liu L., Xiong Q., Flores C., Wong J., Shi J., Wang X., Liu X., Xiang Q., Jiang S. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013;45:330–333. doi: 10.1038/ng.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Shi J., Zhai Y., Hou Y., Ma W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. U S A. 2015;112:5850–5855. doi: 10.1073/pnas.1421475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Guirao R., Baldrich P., Weigel D., Rubio-Somoza I., San Segundo B. The microRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Mol. Plant Microbe Interact. 2018;31:249–259. doi: 10.1094/MPMI-05-17-0108-R. [DOI] [PubMed] [Google Scholar]
- Salvador-Guirao R., Hsing Y.I., San Segundo B. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front. Plant Sci. 2018;9:337. doi: 10.3389/fpls.2018.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Sanuy F., Peris-Peris C., Tomiyama S., Okada K., Hsing Y.I., San Segundo B., Campo S. Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus. BMC Plant Biol. 2019;19:563. doi: 10.1186/s12870-019-2156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.S., Diloknawarit P., Park B.S., Chua N.H. ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol. 2019;221:2067–2079. doi: 10.1111/nph.15530. [DOI] [PubMed] [Google Scholar]
- Seo J.S., Sun H.X., Park B.S., Huang C.H., Yeh S.D., Jung C., Chua N.H. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell. 2017;29:1024–1038. doi: 10.1105/tpc.16.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A., Baulcombe D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D., Molnar A., Lucioli A., Szittya G., Hornyik C., Tavazza M., Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N.A., Eamens A.L., Wang M.B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011;7:e1002022. doi: 10.1371/journal.ppat.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Li Y., Cao X., Qi Y. microRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019;70:489–525. doi: 10.1146/annurev-arplant-050718-100334. [DOI] [PubMed] [Google Scholar]
- Song X., Li P., Zhai J., Zhou M., Ma L., Liu B., Jeong D.H., Nakano M., Cao S., Liu C. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- Song X., Wang D., Ma L., Chen Z., Li P., Cui X., Liu C., Cao S., Chu C., Tao Y. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J. 2012;71:378–389. doi: 10.1111/j.1365-313X.2012.05001.x. [DOI] [PubMed] [Google Scholar]
- Soto-Suarez M., Baldrich P., Weigel D., Rubio-Somoza I., San Segundo B. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci. Rep. 2017;7:44898. doi: 10.1038/srep44898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu P.D. RNA-protein interactions in plant disease: hackers at the dinner table. New Phytol. 2015;207:991–995. doi: 10.1111/nph.13495. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. Network properties of robust immunity in plants. Plos Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Wang H., Jiao X., Kong X., Hamera S., Wu Y., Chen X., Fang R., Yan Y. A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol. 2016;170:2365–2377. doi: 10.1104/pp.15.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Lin F.M., Thomma B.P., Huang H.D., Jin H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants. 2016;2:16151. doi: 10.1038/nplants.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yu W., Yang Y., Li X., Chen T., Liu T., Ma N., Yang X., Liu R., Zhang B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015;5:16946. doi: 10.1038/srep16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Meng X., Dobrovolskaya O.B., Orlov Y.L., Chen M. Non-coding RNAs and their roles in stress response in plants. Genom Proteom Bioinf. 2017;15:301–312. doi: 10.1016/j.gpb.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Liu Y., Li D., Li L., Zhang Q., Wang S., Huang H. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial Canker pathogen invasion. Front. Plant Sci. 2017;8:413. doi: 10.3389/fpls.2017.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Dellota E., Jr., Yamane D., Jin H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017;14:421–428. doi: 10.1080/15476286.2017.1291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xia Y., Lin S., Wang Y., Guo B., Song X., Ding S., Zheng L., Feng R., Chen S. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018;95:584–597. doi: 10.1111/tpj.13972. [DOI] [PubMed] [Google Scholar]
- Wang N., Zhang D., Wang Z., Xun H., Ma J., Wang H., Huang W., Liu Y., Lin X., Li N. Mutation of the RDR1 gene caused genome-wide changes in gene expression, regional variation in small RNA clusters and localized alteration in DNA methylation in rice. BMC Plant Biol. 2014;14:177. doi: 10.1186/1471-2229-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tyler B.M., Wang Y. Defense and counter defense during plant-pathogenic oomycete infection. Annu. Rev. Microbiol. 2019;73:667–696. doi: 10.1146/annurev-micro-020518-120022. [DOI] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.M., Zhao H., Zhang Z., Kaloshian I., Huang H.D., Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Yang R., Yang Z., Yao S., Zhao S., Wang Y., Li P., Song X., Jin L., Zhou T. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants. 2017;3:16203. doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- Wu J., Yang Z., Wang Y., Zheng L., Ye R., Ji Y., Zhao S., Ji S., Liu R., Xu L. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife. 2015;4:e05733. doi: 10.7554/eLife.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhang Q., Zhou H., Ni F., Wu X., Qi Y. Rice microRNA effector complexes and targets. Plant Cell. 2009;21:3421–3435. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Song N., Hu Z., Qin D., Xie C., Peng H., Ni Z., Sun Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Li P., Mei H., Wang D., Sun J., Yang C., Hao L., Cao S., Chu C., Hu S. Fine-tuning of MiR528 accumulation modulates flowering time in rice. Mol. Plant. 2019;12:1103–1113. doi: 10.1016/j.molp.2019.04.009. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu T., Shen D., Wang J., Ling X., Hu Z., Chen T., Hu J., Huang J., Yu W. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathog. 2019;15:e1007534. doi: 10.1371/journal.ppat.1007534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Yang Z., Yang R., Huang Y., Guo G., Kong X., Lan Y., Zhou T., Wang H., Wang W. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant. 2019;12:1114–1122. doi: 10.1016/j.molp.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Ye C.Y., Chen L., Liu C., Zhu Q.H., Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- Yin C., Ramachandran S.R., Zhai Y., Bu C., Pappu H.R., Hulbert S.H. A novel fungal effector from Puccinia graminis suppressing RNA silencing and plant defense responses. New Phytol. 2019;222:1561–1572. doi: 10.1111/nph.15676. [DOI] [PubMed] [Google Scholar]
- Yu Y., Zhou Y.F., Feng Y.Z., He H., Lian J.P., Yang Y.W., Lei M.Q., Zhang Y.C., Chen Y.Q. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol. J. 2020;18:679–690. doi: 10.1111/pbi.13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Jeong D.H., De Paoli E., Park S., Rosen B.D., Li Y., Gonzalez A.J., Yan Z., Kitto S.L., Grusak M.A. microRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ding Z., Wu K., Yang L., Li Y., Yang Z., Shi S., Liu X., Zhao S., Yang Z. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant. 2016;9:1302–1314. doi: 10.1016/j.molp.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Zhang H., Tao Z., Hong H., Chen Z., Wu C., Li X., Xiao J., Wang S. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants. 2016;2:16016. doi: 10.1038/nplants.2016.16. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhao Y.L., Zhao J.H., Wang S., Jin Y., Chen Z.Q., Fang Y.Y., Hua C.L., Ding S.W., Guo H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants. 2016;2:16153. doi: 10.1038/nplants.2016.153. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang M., Li N., Wang H., Qiu P., Pei L., Xu Z., Wang T., Gao E., Liu J. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 2018;16:1172–1185. doi: 10.1111/pbi.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bao Y., Shan D., Wang Z., Song X., Wang Z., Wang J., He L., Wu L., Zhang Z. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018;177:352–368. doi: 10.1104/pp.17.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao H., Gao S., Wang W.C., Katiyar-Agarwal S., Huang H.D., Raikhel N., Jin H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Li J., Lian B., Gu H., Li Y., Qi Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018;9:5056. doi: 10.1038/s41467-018-07500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.M., Zhang Y. Plant immunity: danger perception and signaling. Cell. 2020;181:978–989. doi: 10.1016/j.cell.2020.04.028. [DOI] [PubMed] [Google Scholar]
- Zhu C., Liu T., Chang Y.N., Duan C.G. Small RNA functions as a trafficking effector in plant immunity. Int. J. Mol. Sci. 2019;20:2816. doi: 10.3390/ijms20112816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Wang S., Zhou Y., Bai J., Huang G., Liu X., Zhang Y., Tang D., Lu D. Transcriptional regulation of the immune receptor FLS2 controls the ontogeny of plant innate immunity. Plant Cell. 2018;30:2779–2794. doi: 10.1105/tpc.18.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Wang Q., Zhu B., Luo Y., Gao L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun. 2016;479:132–138. doi: 10.1016/j.bbrc.2016.07.032. [DOI] [PubMed] [Google Scholar]