Figure 1.
NLP1 directly binds and activates CLE35 expression.
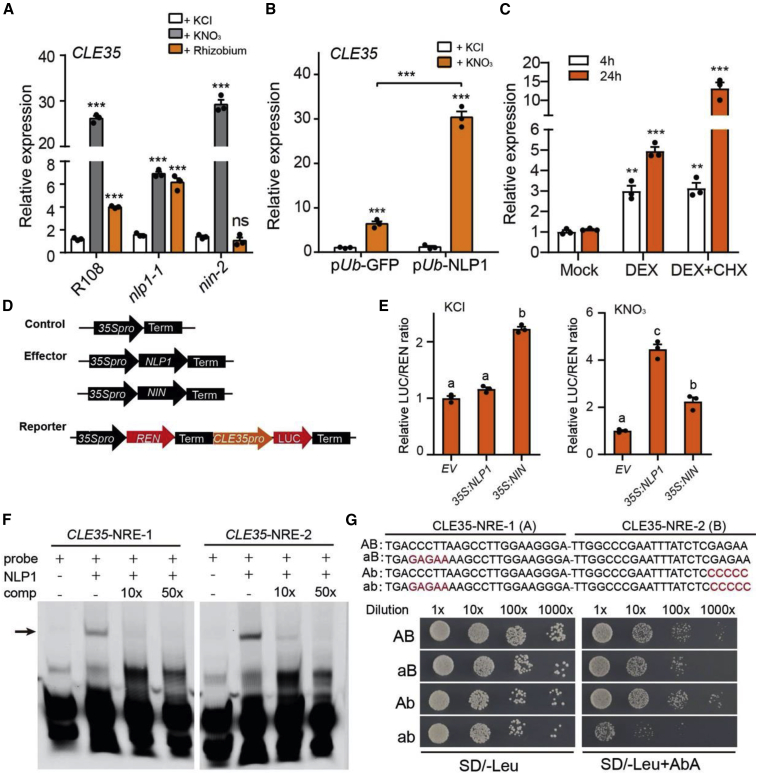
(A) Relative expression of CLE35 in wild-type (R108), nlp1-1, and nin-2 roots in response to nitrate treatment (10 mM, 3 days) or rhizobial inoculation (3 dpi).
(B) Relative expression of CLE35 in EV control (pUb-GFP) and NLP1-overexpressing (pUb-NLP1) hairy roots in response to nitrate (10 mM, 3 days).
(C) Relative expression of CLE35 in pUb-NLP1-GR roots. Roots were incubated in liquid FP medium supplemented with mock (DMSO), dexamethasone (DEX), or DEX + cycloheximide (CHX) for 4 or 24 h. Asterisks indicate significant differences (two-tailed t-test, ∗∗P < 0.01, ∗∗∗P < 0.001).
(D and E) NLP1 and NIN activate CLE35 expression in Nicotiana benthamiana leaves. qRT-PCR was used to measure the gene expression levels in treated roots; expression levels were normalized relative to the value of the ubiquitin reference gene. Data from one representative experiment of three biological replicates are shown. (D) Schematic diagrams of the CLE35 promoter-driven dual-luciferase (LUC) reporter gene and effector genes (control, NLP1, and NIN). (E) The luciferase activity of co-expressed effector gene and CLE35 promoter in N. benthamiana in the absence (KCl) or presence (KNO3) of nitrate. Luciferase activity was normalized to REN activity, and fold changes are relative to the co-expression with EV. Different letters in (E) indicate significant differences from the respective controls based on ANOVA with a post hoc Tukey's test (P < 0.05). Error bars represent mean ± SEM.
(F) Gel-shift assays of NLP1 binding to the promoter of CLE35. The protein–DNA complexes were separated by electrophoresis, and the fluorescence-labeled DNA was detected by fluorimetry. Unlabeled DNA fragments were added as competitors (comp). Arrow indicates the retardation of migration of the NLP1 bound to the probes.
(G) Yeast one-hybrid assay of NLP1 binding to the two NRE regions in the CLE35 promoter. Binding assays were performed for NLP1 against the synthetic NRE sequence (AB) or the mutated NRE sequence (aB, Ab, and ab, in red). The medium lacking leucine (−Leu) was supplemented with 250 ng/ml aureobasidin A (AbA). Yeast cells were diluted in a 10× dilution series.