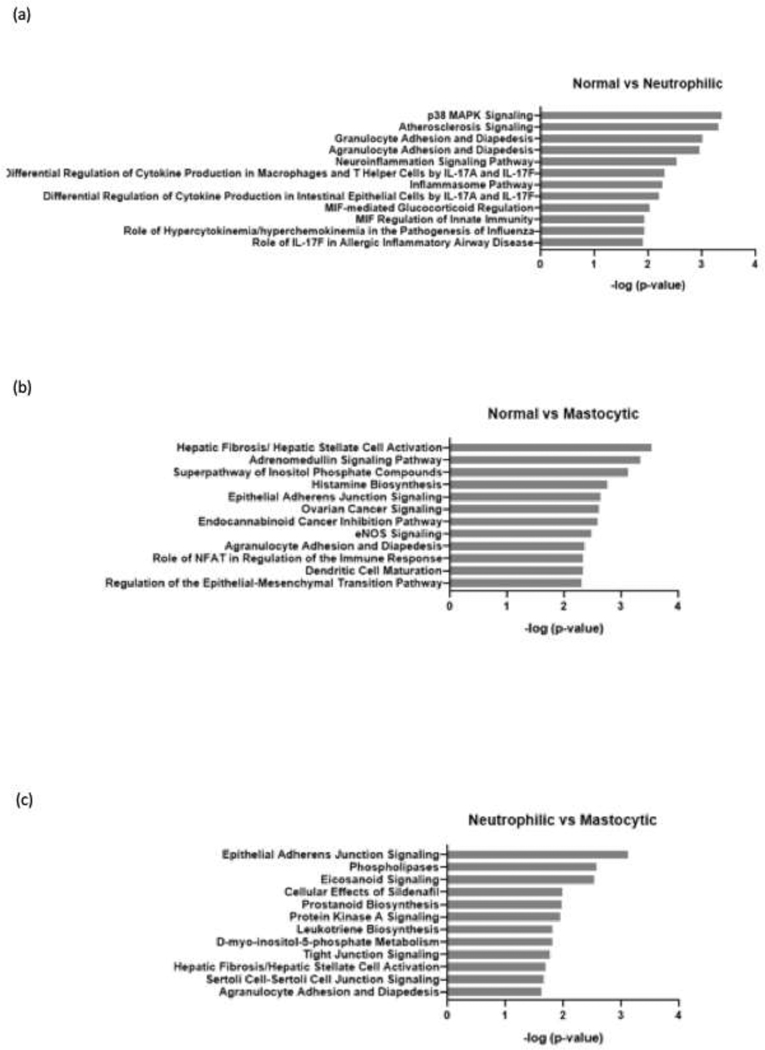
Figure 5.
Summary of pathway analysis for differentially expressed genes. The differentially expressed genes were compared between normal vs. neutrophilic phenotype (a), normal vs. masotytic phenotype (b), and neutrophilic vs. mastocytic phenotype (c). Functional annotation was performed using Ingenuity Pathway Analysis using predefined pathways and functional categories of the Ingenuity Knowledge Base. Fisher’s exact test was applied to identify significantly enriched differentially expressed genes as members of pathways and functional categories. Relevant gene regulatory networks were identified using the Ingenuity Knowledge Base.