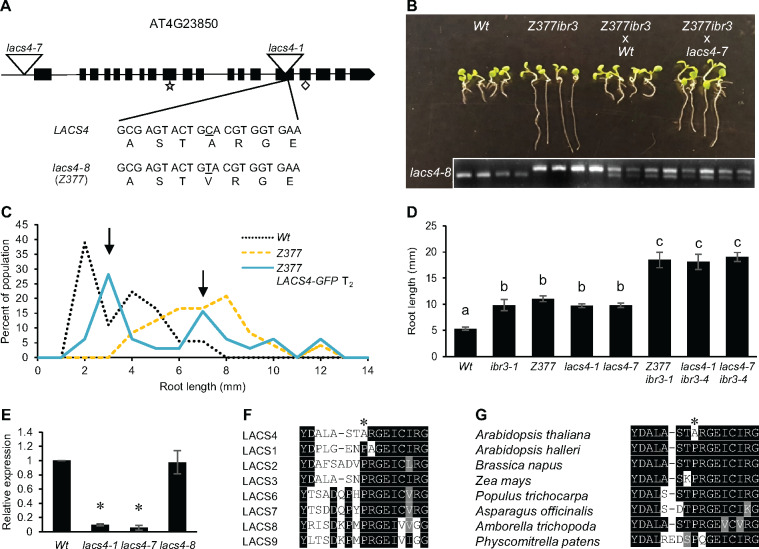
Figure 2.
Z377 is defective in lacs4. (A) Schematic of LACS4 gene structure. Exons are represented as black boxes whereas introns are lines. Locations of T-DNA insertion mutants used in this study are depicted as inverted triangles. The mutation in Z377 changes nucleotide 1,400 from C to T, resulting in an alanine to valine substitution at residue 467. As a result, Z377 was renamed as lacs4-8. Predicted AMP and fatty acid binding motifs (UniProt Consortium, 2019) are indicated by a star and diamond, respectively. (B) Z377 ibr3-1 was crossed to Wt and lacs4-7 in a test of non-complementation. F1 seedlings were grown on 20 µM IBA for 7 d. Seedlings were genotyped for the point mutation in LACS4 in Z377 to confirm the cross was successful. The Wt LACS4 allele is the smaller product whereas the mutant lacs4 is the larger product. (C) Z377 seedlings segregating a Wt copy of LACS4 tagged with a C-terminal GFP were grown on 15 µM IBA for 7 d. Length of primary root was measured for each individual. Data are represented as percent of the population with indicated primary root length (n ≥ 18 Wt and Z377, n = 32 Z377 35S: LACS4-GFP). (D) Primary root elongation of 7-d-old seedlings grown on 20 µM IBA. Statistical significance was determined by one-way ANOVA with post hoc Tukey HSD test (±SE, n ≥ 11, P < 0.05). Common letters indicate no significant difference. (E) Expression of LACS4 relative to Wt in 5-d-old seedlings grown under white light on filter paper. LACS4 expression is normalized against UBQ10. Statistical significance was determined by a two-tailed t test (± se, n = 5, *P < 0.001). (F) and (G) T-Coffee alignment of LACS proteins of A. thaliana (F) and LACS4 orthologs in divergent plant species (G). Residues depicted are 460–475 of LACS4 from A. thaliana. The amino acid residue mutated in lacs4-8 is indicated with an asterisk. Black shading indicates the same amino acid. Gray shading indicates similar amino acids.