Figure 1.
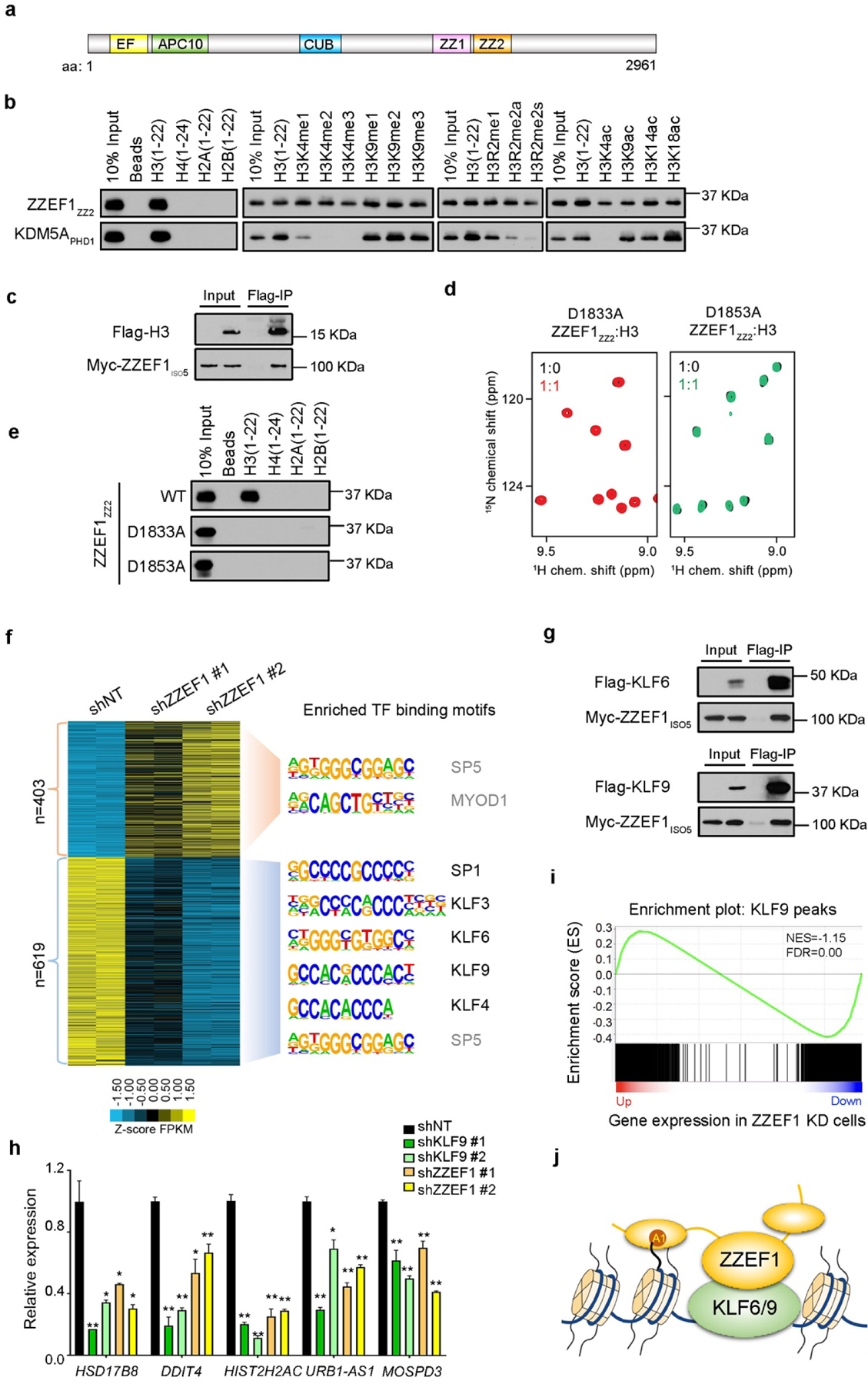
ZZEF1 binds to histone H3 and the KLF6/9 transcription factors to stimulate gene expression. a, Schematic representation of ZZEF1 protein structure. EF: EF-hand motif; APC10: APC10/DOC1-like domain; CUB; CUB domain; ZZ1: the first ZZ domain; ZZ2: the second ZZ domain. b, Western blot analysis of peptide pull-downs of GST-tagged ZZEF1ZZ2 and KDM5APHD1 with the indicated histone peptides. c, Western blot analysis of Flag co-IP in cells expressing H3-Flag and Myc- ZZEF1iso5. d, Superimposed 1H, 15N HSQC spectra of the indicated ZZEF1ZZ2 mutants titrated with the H3(1–12) peptide. e, Western blot analysis of peptides pull-downs of WT ZZEF1ZZ2 and the indicated mutants with histone peptides. f, Heatmap of upregulated (n=403) and downregulated (n=619) genes in ZZEF1 shRNA knockdown (KD) cells compared with control (shNT) cells. Enriched consensus motifs and transcription factors (TFs) are shown on the right. TFs that are not expressed (RPKM<1) in H1299 cells are shown in gray. g, Western blot analysis of Flag co-IP in cells expressing Flag-KLF6 or KLF9 and Myc-ZZEF1iso5. h, qRT-PCR analysis of gene expressions in control (shNT) and KLF9 or ZZEF1 KD H1299 cells. Error bars indicate SD of three replicates. *: p<0.05; **: p<0.01 (Student’s t-test) i, GSEA analysis showing that down-regulated genes in ZZEF1 KD cells are enriched for KLF9 occupancy. NES: normalized enrichment score, FDR: false discovery rate. j, Working model of ZZEF1 binding to H3 and KLF6/9 to promote gene expression. A1: Ala 1 of histone H3.
