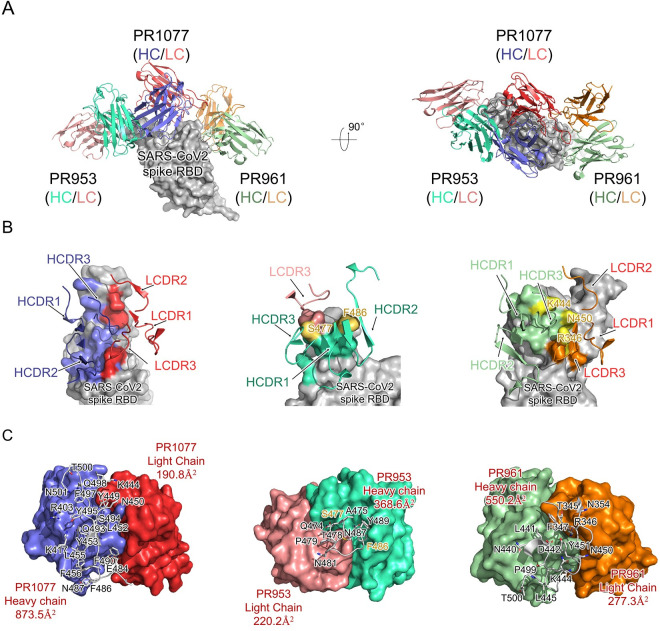
Fig 3. Structural analysis of the SARS-CoV-2 RBD-NAbs scFv complex.
(A) Overall structures of the PR1077, PR953, and PR961-Fab-RBD complexes. The PR1077 HC (colored slate blue) and light chain (colored red) are shown. PR953 heavy (colored green cyan) and light (colored salmon red) chains and PR961 heavy (colored pale green) and light (colored orange) chains are displayed. The SARS-CoV-2 RBD is colored in gray and displayed in surface representation. (B) The epitopes of antibodies are shown in surface representation. The CDR loops of NAbs are colored as above. The S477 and F486 residues in contact with both the heavy and light chains of PR953 are colored in yellow; 3 hydrogen donor residues (R346, K444, and N450) which contact with both the heavy and light chains of PR961 are colored in yellow. (C) The binding surface of NAbs with SARS-CoV-2-RBD. The interaction residues of RBD are shown in sticks and labeled accordingly; variable regions’ interaction surfaces of NAbs are colored as above. HC, heavy chain; HCDR, heavy chain complementarity-determining region; LCDR, light chain complementary-determining region; NAb, neutralizing antibody; RBD, receptor-binding domain; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; scFv, single-chain variable fragment.