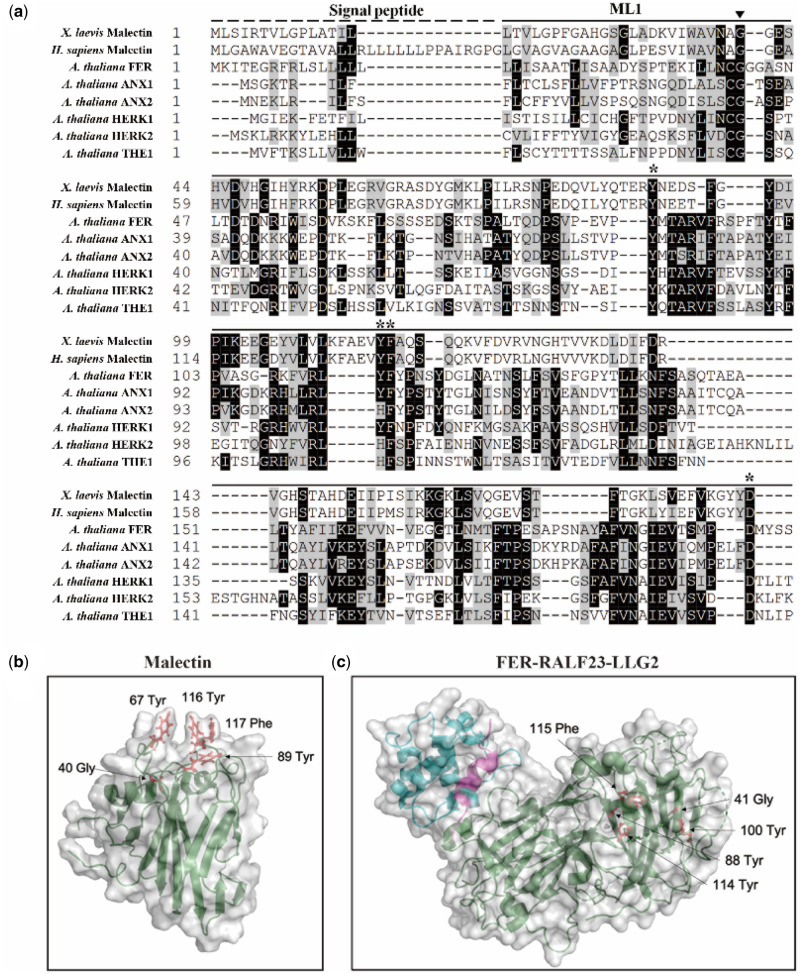
Figure 5.
Localization of the highly conserved glycine mutation in fer-ts in Arabidopsis FERONIA:RALF23:LLG2 and X. laevis malectin structures. (A) Several residues important for binding carbohydrate ligands are conserved in plant CrRLK1L receptor kinase family members. Sequences analyzed include animal Malectin (X. laevis and Homo sapiens) FERONIA and other well-characterized CrRLK1L homologs in Arabidopsis (ANXUR1; ANX1, ANXUR1; ANX2, HERCULES1; HERK1 and THESEUS1; THE1). Putative N-terminal SPs are indicated as black dashed lines and malectin and CrRLK1L ML1 domains by solid lines, respectively. The highly conserved G41 of FER is marked by an arrowhead. Black boxes indicate fully conserved residues; shaded boxes indicate similar and partially conserved residues. Conserved residues that have been shown to participate in binding nigerose in the X. laevis malectin structure are marked by asterisks. Sequence alignment analysis was performed by CLUSTAL Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) and displayed by using BOXSHADE software (www.ch.embnet.org/software/BOX_form.html). (B) Crystal structure of the X. laevis malectin protein (PDB ID: 2K46) with binding pocket aromatic residues and the highly conserved glycine residue based on sequence similarity to FER shown in red. C, Crystal structure of FER protein (green) in complex with RALF23 ligand (magenta) and glycosylphosphatidylinositol-anchored protein LLG2 (blue) (PDB ID: 6A5E). No analogous binding pocket is observed on the ML1 domain, as all conserved aromatic residues (red) are buried within the protein. Both (B and C) were generated using PyMol (DeLano Scientific).