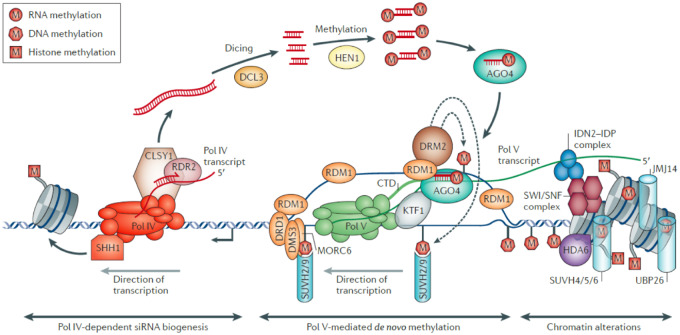
Figure 3.
Establishment of post transcriptional silencing, RNA-directed methylation, and eventually stable transcriptional silencing of transposons. Left panel, Transcript of new active transposon by RNA Polymerase II is made double stranded by RNA-directed RNA Polymerase 6 and the double stranded RNA is then cleaved into 21 and 22 nt pieces by Dicers 2 and 4. These pieces become loaded on to Argonaute (AGO) 1 and guide cleavage of the RNA transcripts of the transposon to achieve post transcriptional silencing (PTGS) (Panda and Slotkin, 2013). Middle Panel, The 21–22 nt sRNAs can also trigger some methylation of the transposon by associating via base pairing with a Polymerase V transcript of the transposon. This complex attracts the DOMAINS REARRANGED METHYTRANSFERASE 2 (DRM2) which achieves some methylation of the transposon DNA and transcription gene silencing (TGS). Right panel, Polymerase IV can be attracted to the methylated transposon DNA and the transposon transcript can be made double stranded by RNA-dependent Polymerase 2 and cleaved with Dicer 3 into 24 nt pieces. Following incorporation into Argonaute 4 the 24 nt RNA can base pair with a Polymerase V transcript which stimulates DRM2 recruitment and dense methylation of the DNA. Once the transposon is substantially methylated then 24 nt RNAs are produced by the Polymerase IV pathway (Middle panel) to ensure continued dense methylation and transcriptional silencing. Reprinted from Matzke MA and Mosher RA, (2014) with permission from Nature/Springer.