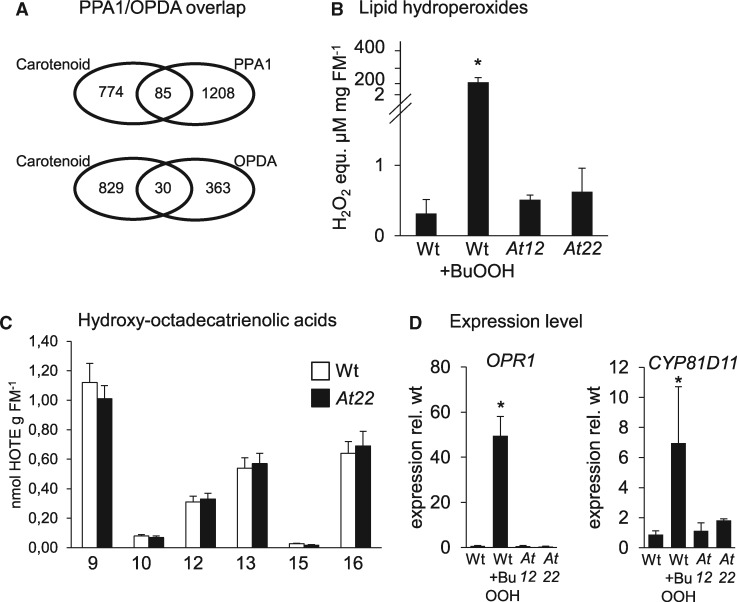
Figure 5.
Lipid hydroperoxides in Arabidoposis roots. (A) Venn diagrams showing the overlap between commonly regulated DEGs in roots of PSY-overexpressing lines At12 and At22 (Carotenoid) and genes regulated by treatment with the oxylipins PPA1 and OPDA, respectively. (B) Total lipid hydroperoxides in roots from wild type, At12 and At22. WT was treated with 100-mM BuOOH for 4 h to induce lipid peroxidation. Lipid hydroperoxide levels were determined with the FOX2 assay and are given in H2O2 equivalents per mg fresh mass (FM). Data are mean ± sd from three biological replicates. Significant difference, Student’s t test, *P <0.05. (C) HOTE isomer pattern in Arabidopsis roots. The number indicates the position of the hydroxy group. 10- and 15-HOTEs are specifically formed by 1O2 and accordingly low in (non-photosynthetic) roots while 9-, 12-, 13-, and 16-HOTEs are generated through both 1O2 and free radical catalysis. Similar HOTE patterns in WT and carotenoid-accumulating roots (line At22) indicate that lipid oxidation can be excluded in transcriptome responses observed. Data are means ± sem from nine biological replicates each. HOTEs are given in nmol g-1 fresh mass (FM). (D) Expression level of lipid stress marker genes for OPR1 and the cytochrome p450 enzyme CYP81D11. Transcripts were normalized to 18S rRNA levels and are expressed relative to levels in untreated WT. Data are mean ± sd from three biological replicates. Significant difference, Student’s t test, *P <0.05.