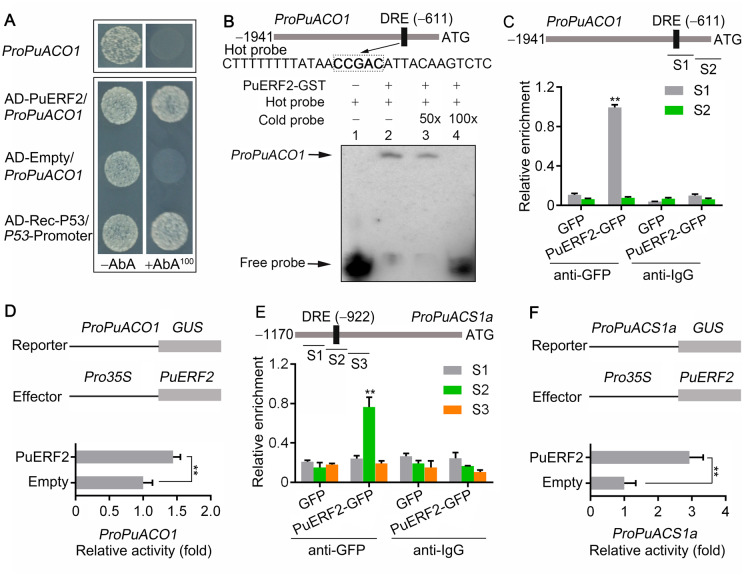
Figure 5.
PuERF2 regulates PuACO1 and PuACS1a transcription. A, Y1H analysis showing that PuERF2 binds to the promoter of PuACO1 (ProPuACO1). See Figure 4A for additional details. B, EMSA showing that PuERF2 binds to the DRE motif of the PuACO1 promoter. The hot probe was a biotin-labeled fragment of the PuACO1 promoter containing the DRE motif, and the cold probe was a non-labeled competitive probe (50- and 100-fold that of the hot probe). GST-tagged PuERF2 was purified. C, ChIP-PCR showing the in vivo binding of PuERF2 to the PuACO1 promoter. Cross-linked chromatin samples were extracted from PuERF2-GFP overexpressing fruit calli (PuERF2-GFP) and precipitated with an anti-GFP antibody. Eluted DNA was used to amplify the sequences neighboring the DRE motif by qPCR. Two regions (S1 and S2) were investigated. Fruit calli overexpressing the GFP sequence (GFP) were used as negative controls. Values are the percentage of DNA fragments that co-immunoprecipitated with the GFP antibody or a nonspecific antibody (pre-immune rabbit IgG) relative to the input DNA. The ChIP assay was repeated 3 times and the enriched DNA fragments in each ChIP were used as one biological replicate for qPCR. Values represent means ± se. Statistical significance was determined using a Student’s t test (**P < 0.01). D, GUS activity analysis showing that PuERF2 promotes the activity of the PuACO1 promoter. The PuERF2 effector vector and the reporter vector containing the PuACO1 promoter were infiltrated into N. benthamiana leaves to analyze the regulation of GUS activity. Three independent transfection experiments were performed. Values represent means ± se. Statistical significance was determined using a Student’s t test (**P < 0.01). E, ChIP-PCR showing the in vivo binding of PuERF2 to the PuACS1a promoter. F, GUS activity analysis showing that PuERF2 promotes the PuACS1a promoter.