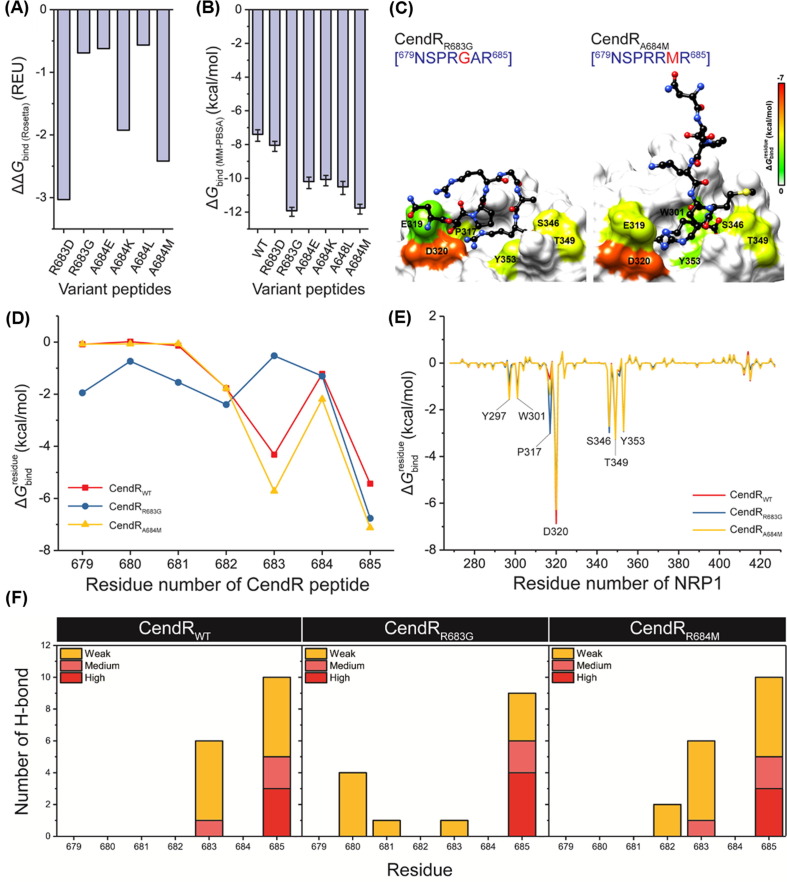
Fig. 7.
(A) ΔΔGbind(Rosetta) (REU) of all designed peptides obtained from Rosetta. Note that the ΔΔGbind was calculated using the following equation: ΔΔGbind = ΔGbind (mutant) – ΔGbind (wild-type). (B) ΔGbind (kcal/mol) of all designed peptides obtained from MM-PBSA method. (C) Representative structures of the two most promising peptides CendRR683G and CendRA684M showing the ligand orientation in NRP1 b1 domain drawn from the last MD snapshot. (D) of CendRWT, CendRR683G, and CendRA684M contributing to the binding of NRP1. (E) of NRP1 contributing to the binding of CendRWT, CendRR683G, and CendRA684M. (F) Percentage of H-bond occupation of NRP1 contributing to the binding of (left) CendRWT, (middle) CendRR683G, and (right) CendRA684M. %H-bond occupation (%H-bondoc) was classified into four levels: (i) strong H-bond (%H-bondoc of > 75%), (ii) medium H-bond (75% ≥ %H-bondoc > 50%), and (iii) weak H-bond (50% ≥ %H-bondoc > 10%).