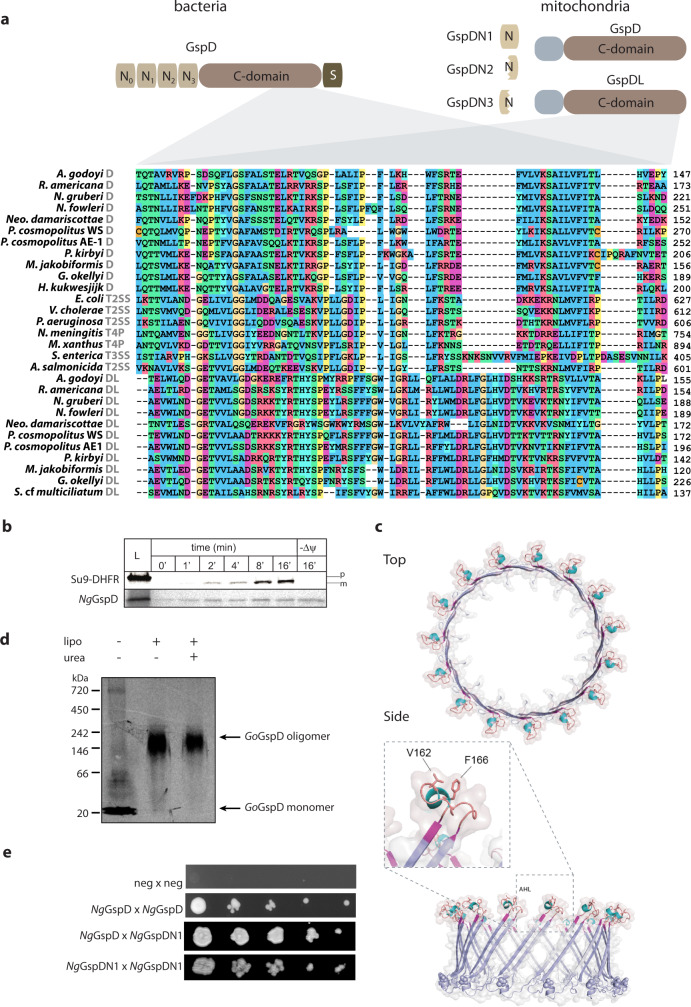
Fig. 4. Mitochondrial GspD oligomerizes towards the formation of membrane pores.
a Domain architecture of the canonical bacterial GspD and the eukaryotic proteins homologous to its different parts (short N-terminal region of mitochondrial GspD of unidentified homology shown in grey). Below, protein sequence alignment of the secretin C-domain of bacterial and mitochondrial orthologues (mitochondrial GspD or GspDL and the respective molecular complex of bacterial secretins is depicted in grey, the numbers on the right depict the position of the amino acid in the particular sequence). b In vitro import of NgGspD into isolated yeast mitochondria over a period of 16 min. Dissipation of the membrane potential (ΔΨ) by AVO mix abolished the import of matrix reporter protein (Su9-DHFR) bud did not affect the mitochondrial GspD; p precursor of Su9-DHFR, m mature form of the protein upon cleavage of the mitochondrial targeting sequence. c Structural model of GoGspD built by ProMod3 on the Vibrio cholerae GspD template. Top and side view of a cartoon and a transparent surface representation of the GoGspD pentadecamer model is shown in blue. The amphipathic helical loop (AHL), a signature of the secretin family, is highlighted and coloured according to the secondary structure with strands in magenta, helices in cyan and loops in light brown. The C-terminal GpsD residues are highlighted as spheres. The detailed view of the AHL region shows the essential residues V162 and F166 pointing towards the membrane surface. d In vitro translation and assembly of mitochondrial GoGspD into a high-molecular-weight complex; lipo liposomes added, urea liposome fraction after 2 M urea treatment. e Y2H assay suggests the self- and mutual interaction of NgGspD and NgGspDN1. (for b–d, representative images of three experiments are shown).