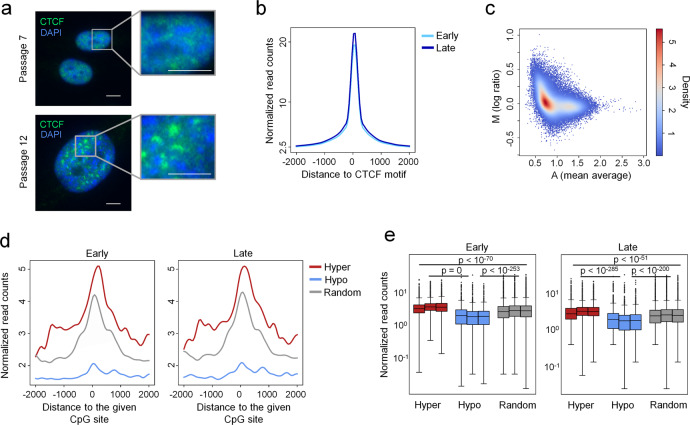
Fig. 6. CTCF binding in cells of early and late passages.
a High-resolution microscopy pictures of MSCs at median passage (P7) and late passage (P12). Staining of cell nuclei with DAPI and CTCF reveals co-localization of CTCF particularly in the larger nuclei of late passage MSCs (size bars = 5 µm). b CTCF ChIP-seq signals of cells in early (P2) and late (P8–P14) passage are centered around predicted CTCF binding motifs. c MA plot of CTCF ChIP-seq signals of cells in early (P2) and late (P8–P14) passage. Mean average (A) is the mean of the normalized read counts of early and late passages, whereas log ratio (M) is the ratio of the normalized read counts of late passages over early passages. d Lineplots depict the CTCF ChIP-seq signals of cells in early (P2) and late (P8–P14) passage centered around hyper- or hypomethylated culture-associated CpGs or 2000 randomly picked CpGs of the Illumina 450k BeadChip. e Statistical testing of the ChIP-seq signal differences of three donors in early (P2) and late (P8–P14) passages at the CpG subsets shown in (d). CTCF ChIP-seq signals at hypermethylated CpGs are significantly enriched in comparison to randomly chosen CpGs, while CTCF ChIP-seq signals at hypomethylated sites are significantly depleted (Dunn’s test on the quantile normalized reads of the 2000 bp window). Raw data of the CTCF ChIP-seq experiment was uploaded to Gene Expression Omnibus (GEO): GSE144196.