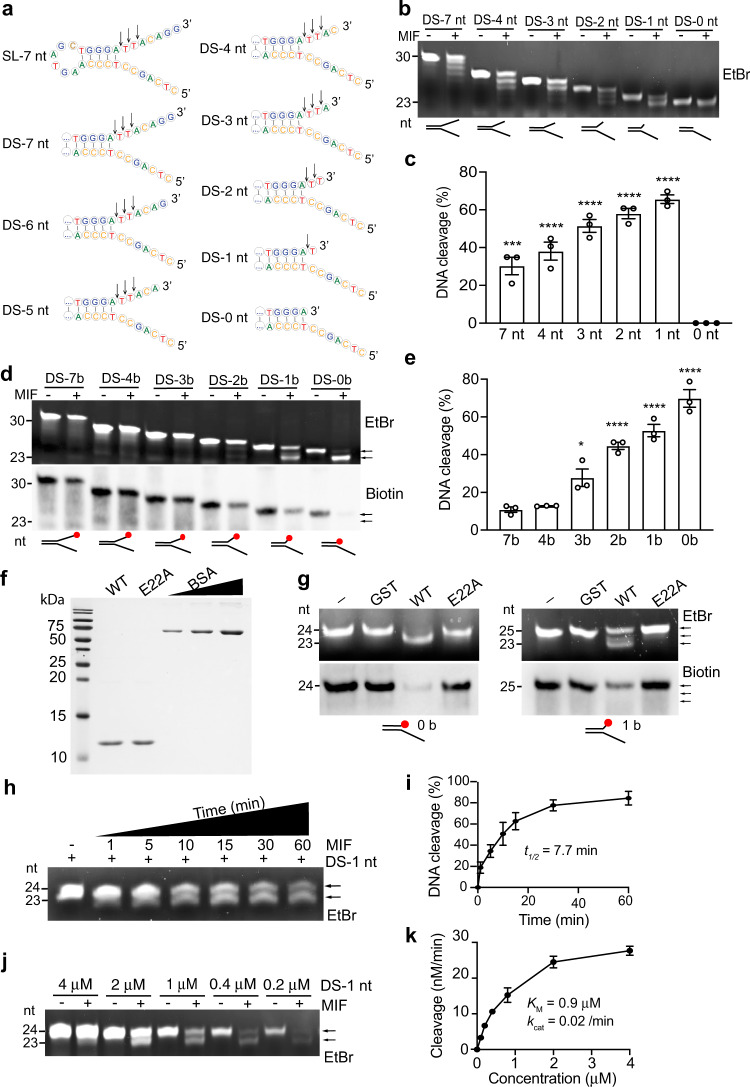
Fig. 1. MIF recognizes Y-shaped dsDNA and cleaves 3’ unpaired nucleotides.
a Second structure of MIF substrates ssDNA with the stem-loop (SL) and Y-shaped dsDNA with different lengths of unpaired nucleotides at the 3′ end. b, c In vitro MIF (2 μM) nuclease cleavage assay using Y-shaped dsDNA as substrates. Data were quantified in c (mean ± SEM, n = 3 biologically independent experiments). ****P < 0.0001, ***P < 0.001; by one-way ANOVA Dunnett’s multiple comparisons test vs 0 nt group. EtBr ethidium bromide. d, e In vitro MIF (2 μM) nuclease cleavage assay using 3′ biotin-labeled Y-shaped dsDNA as substrates. Data were quantified in e (mean ± SEM, n = 3 biologically independent experiments). ****P < 0.0001, *P < 0.05; by one-way ANOVA Dunnett’s multiple comparisons test vs 7b group. Red dot indicates biotin (d). Arrow indicates the cleavage products. f Protein purification of wild-type (WT) MIF and nuclease-inactive E22A mutant shown by Coomassie blue staining. BSA (0.5–3 μg) was used as control. g In vitro nuclease assay of MIF and E22A mutant (2 μM) using 3′ biotin-labeled Y-shaped dsDNA (0.8 μM) as substrates. h, i MIF (2 μM) cleaves DS-1 nt substrate (0.8 μM) in a time (1, 5, 10, 15, 30, 60 min)-dependent manner. Representative image is shown in h. Data are quantified in i (mean ± SEM, n = 3 biologically independent experiments). j, k MIF (2 μM) cleaves DS-1 nt substrate in a concentration (0.2, 0.4, 1, 2, 4 μM)-dependent manner. Representative image is shown in j. Data are quantified in k (mean ± SEM, n = 3 biologically independent experiments).