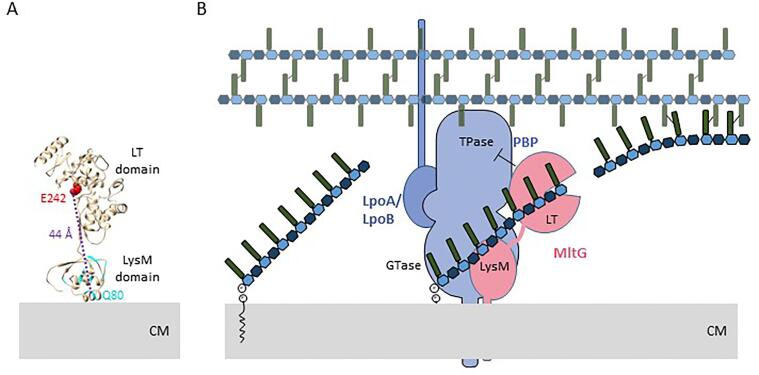
Fig. 5.
Model of glycan strand cleavage by MltG. (A) BsMltG residues were modelled using the crystal structure of Lmo1499 (PDB: 4IIW) as template (Yang et al., 2020). Highly conserved residues in the LysM domain are in cyan (Fig. S1), and the catalytic E242 residue is in red. The distance from the LT catalytic residue E242 to one of the distant and conserved LysM domain residues, Q80, is ~ 44 Å. (B) MltG is recruited to PG synthesis sites, presumably by interacting with PBPs, and competes with their TPase activity, cleaving in the nascent glycan strands to produce strands with 7 disaccharide units. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)