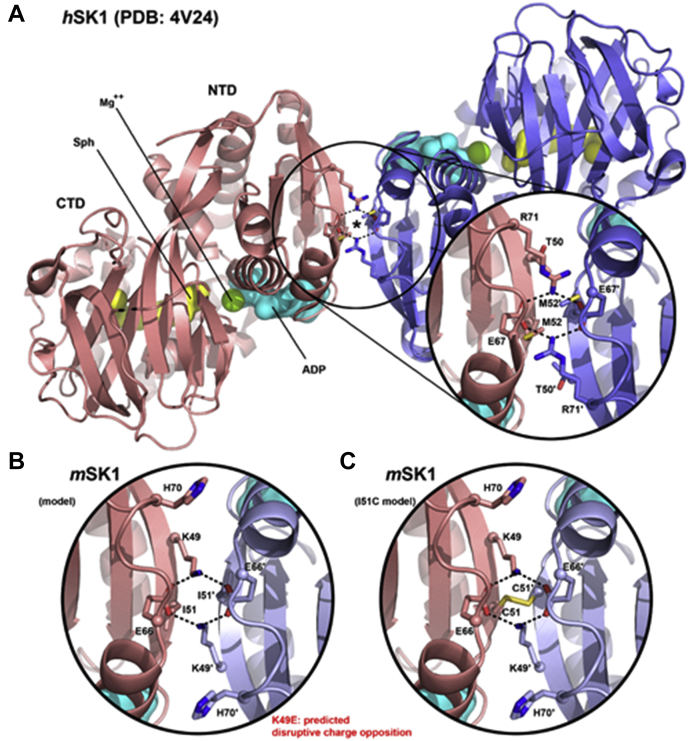
Figure 6.
SK1 dimerization interface and rationale for site-directed mutagenesis.A, crystal structures of hSK1 exhibit head-to-head dimerization with partial annealing of an exposed strand edge (β2) from each protomer about a C2-symmetry axis. A key salt bridge network comprising E67 and R71 in hSK1 contributes to the dimerization interface, as illustrated here from a 1.8 Å resolution structure (PDB ID: 4V24) (21) with the dimerization symmetry axis marked by an asterisk. The positions of bound Sph (yellow surface) and Mg-ADP (green sphere/cyan surface) are shown superimposed from separate crystal structures, 4VZB/4VZD (19). Packing of alternating hydrophobic residues (51-LML-53 in hSK1) along the annealed β2-strand edges also contributes significantly to the dimerization interface (M52 shown here as a stick). B, in mSK1, the glutamic acid of the dimerization salt bridge network is conserved (as E66), but the arginine is replaced by histidine (H70). The additional replacement of a threonine (T50) in hSK1 by a lysine (K49) in mSK1, however, suggests the existence a surrogate E66:K49:E66ʹ:K49ʹ salt bridge network to maintain the dimerization interface in mSK1, as illustrated here (mSK1 homology model). Coincident replacement of M52 in hSK1 with a shorter, branched side chain residue in mSK1 (I51, marked) creates space for the lysines and is also consistent with adoption of the postulated surrogate salt bridge network. Based on this model, we surmised that K49E charge reversal mutation would disrupt the salt bridge network (black dotted lines) to destabilize the dimerization interface. C, the mSK1 homology model of panel B indicated that the Cγ centers for the I51 and I51ʹ residues of the dimer likely lie within van der Waals contact of one another, thereby suggesting a reciprocal strategy to stabilize the dimerization interface by means of an engineered disulfide bridge (as modeled here) through the introduction of I51C substitution. hSK1, human SK1; mSK1, mouse SK1; Sph, sphingosine.